
| Attachments | |
|---|---|
| 7RIS_Ho.01_2.png | 512K |
| 7RIS_autoSHARP_2.png | 16K |
| 7RIS_autoSHARP_1.png | 39K |
| 7RIS_autoSHARP_4.png | 1MB |
| 7RIS_Ho.01_1.png | 224K |
| autosharp_reference_card.pdf | 218K |
| 7RIS_Ca.01_2.png | 515K |
| 7RIS_Ca.01_1.png | 221K |
| 7RIS_autoSHARP_3.png | 898K |
These notes here are specifc for the RapiData 2023 workshop at SSRL. We assume that you know how to (1) connect to NX/NoMachine, (2) start a terminal on one of the processing machines and (3) have some basic familiarity with the command-line.
If you opened a terminal on one of the processing machines, all our software should be setup correctly. Otherwise you might need to run
module load ccp4 module load xds module load autoproc
in order to have a correct setup for our software. In either case, you should now be able to see the online help via
process -hA PDF with the most important (or commonly used) command-line arguments is available here.
If you don't have your own data available for running autoPROC, you can have a look at some example data (mostly data deposited with PDB entries by various users): these are in
/data/rapidata2/SampleData2023
with a separate directory for each project (PDB identifier). In most you will find a subdirectory Images (if the raw diffraction data is available).
There are two datasets available for this PDB entry:
For the tutorial it would be best if you created a fresh directory - e.g. via
mkdir -p ~/rapidata2023/7RISand then go there:
cd ~/rapidata2023/7RIS
Make sure you are on one of the fast processing nodes, i.e.
hostnamereturns something containing pxprocNN (with NN being the node number you are on).
We can just run
process -I /data/rapidata2/SampleData2023/7ris/Images/Ca -d Ca.01 | tee Ca.01.lis
Here
You will now see a lot of text in your terminal, which can be very useful to read since it contains a lot of information. However, it passes quite quickly and scrolling up and down can be tedious. No problem, since we also saved the output into a file (via that tee command above) - so nothing is lost.
The better way of looking at autoPROC results is the so-called "summary.html" file it generates automatically, and which can be found inside the directory specified by the -d flag. So if you followed the suggestions above, you should find it at ~/rapidata2023/7RIS/Ca.01/summary.html and you can open it e.g. via the command
firefox ~/rapidata2023/7RIS/Ca.01/summary.html
Remember to hit the "Reload" button (or Ctrl-R) from time to time: that file is being written to by the currently running process and we might want to refresh what we see. As soon as you see a proper menu on the left hand side (in that slightly grey panel) the job will be finished ... hopefully without any error.
The information in the HTML document should be fairly clear: you will see notes, sometimes warnings and various graphs with explanations (and there are links to only documentation available as well). Some sections are "hidden" by default: symbolised by the little green box with a "+" sign: clicking on it will expand that section and provide you with more information and plots.
The overall sequence of autoPROC steps are
At the end of this autoPROC job you should have quite nice data - e.g. visible via the STARANISO analysis
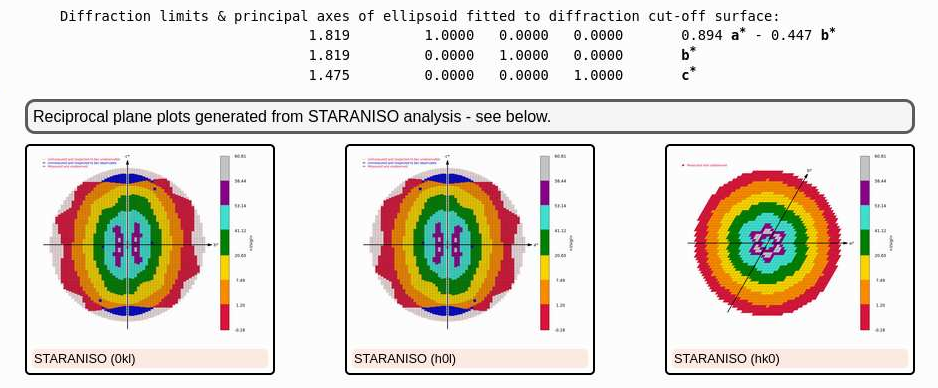 |
and the final set of data-quality metrics
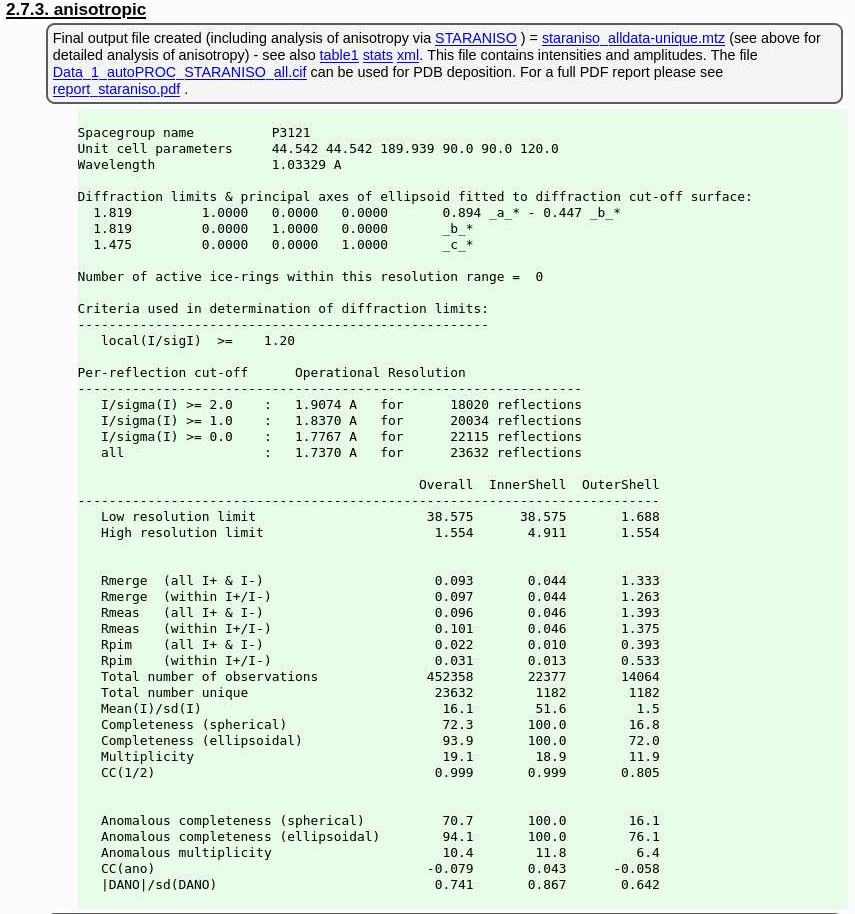 |
We could now process the Holmium dataset in the same way - and we should do so if we didn't know anything else about it. But let's assume that this is just a replacement of the calcium ion and that the crystal form stays basically the same. Then we could use the just processed data as a reference in order to
So let's run
process \ -I /data/rapidata2/SampleData2023/7ris/Images/Ho \ -ref Ca.01/staraniso_alldata-unique.mtz \ -d Ho.01 | tee Ho.01.lis
For better readability we used a backslash (and then a newline) - standard shell syntax to signal continuation (you can still cut-n-paste the above into a terminal as-is).
Now we get an idea about the quality of that dataset:
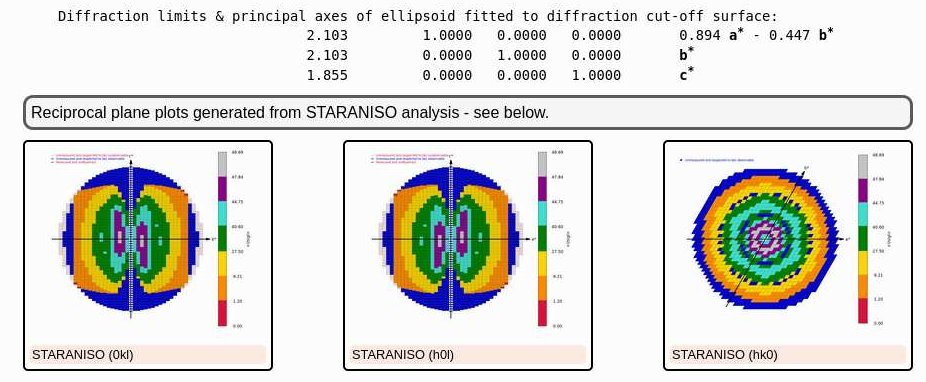 |
(because of the longer wavelength and the long c-axis, the detector could not be moved closer to record the higher diffraction data - as was possible for the Calcium dataset) and the final set of data-quality metrics
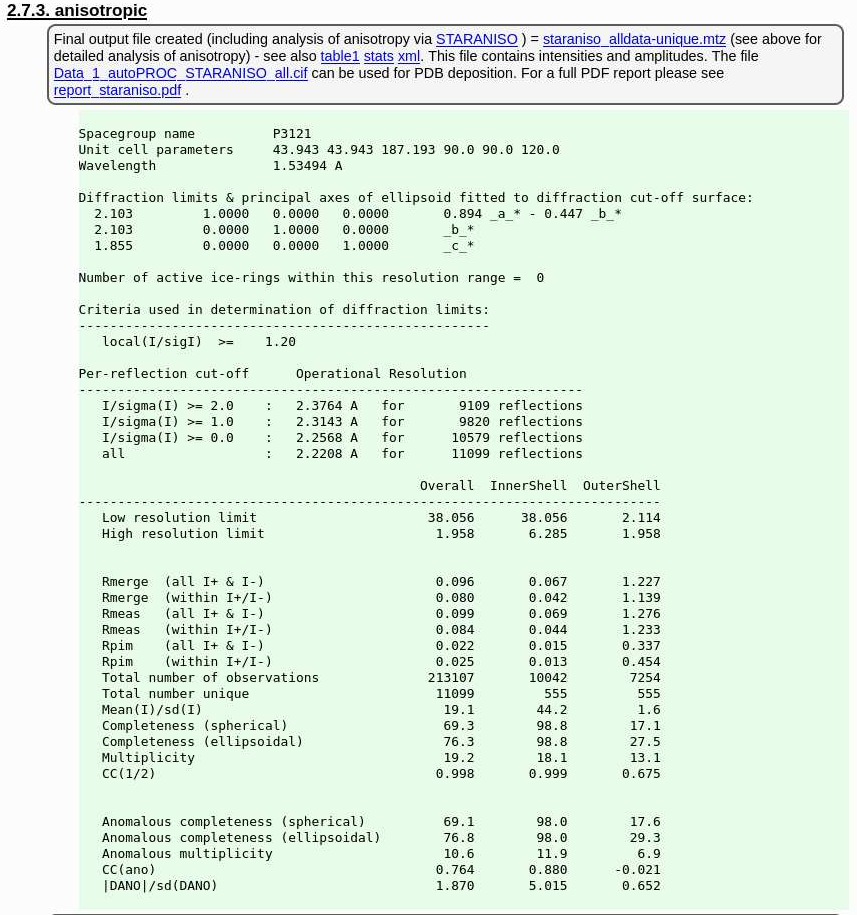 |
Because we have (strong) anomalous signal in the Holmium dataset, we can run SAD phasing. First we need to get the sequence file via
wget -O 7ris.seq https://www.rcsb.org/fasta/entry/7RIS
than we can run
run_autoSHARP.sh \ -seq 7ris.seq \ -ha "Ho" -nsit 1 \ -wvl 1.53494 peak \ -mtz Ho.01/staraniso_alldata-unique.mtz \ -nowarp \ -d autoSHARP.01 | tee autoSHARP.01.lis
The above is using nearly all defaults for SAD phasing (see also run_autoSHARP.sh -h or the PDF cheat-sheet) - apart from the -nowarp flag: this is only to skip the final ARP/wARP model building step to get faster to an already sensible result.
While the job is running you could load the HTML it references (on standard output) - or wait till it is finished. You should then see something like:
 |
and several instructions/suggestions like
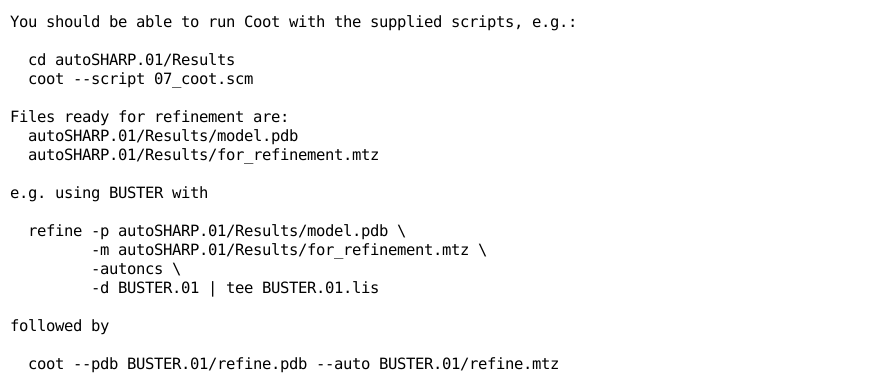 |
Following one of those, namely
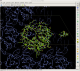
cd autoSHARP.01/Results coot --script 07_coot.scm
allows you to check the current status of the structure solution and initial model building:
 |
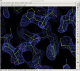 |
That model might not yet be in a state for going directly into refinement (it is after all a quick initial build only), but it shows that the structure can be solved by SAD phasing (as it was done originally) and that the initial model should provide a good starting point for completion - either manually or via some other tools/programs.