EditTNT
|
Copyright © 2004-2008 Global Phasing Limited
All rights reserved.
This software is proprietary to and embodies the
confidential technology of Global Phasing Limited
(GPhL).
Possession, use, duplication or dissemination of
the software is
authorised only pursuant to a valid
written licence from GPhL.
Author: (2004-2008) M. Brandl
|
Contents
EditTNT is a Java-based interactive tool for editing and visualization
of TNT-restraints. All types of restraints
are supported. You can modify existing restraints or delete them and
add new ones. Visualization is carried out by JMOL.
Provided with a PDB-file (see below: How to start EditTNT), EditTNT
can call Gelly (O.Smart) to optimize the PDB-file with the parameters chosen (including
the inversion of chiralities by sign change of the corresponding improper torsion angles).
EditTNT can be used to visualize restraints in PDB, MDL and Tripos-MOL2-files
|
% EditTNT [TNT-dictionary-file] [structure-file]
|
where:
| Argument |
Description |
Note |
| TNT-dictionary-file |
TNT-style-restraints-file |
|
| structure-file |
name of PDB, MDL, or Tripos Mol2-file |
matches up with the TNT-dictionary file |
See also
for a list of command line arguments. For examples, please refer to
the Tutorial section of this documentation.
- The main table
- Click on the tab BOND, ANGLE,
TORSION, etc. to view a table of restraints of the
given kind
- Use the mouse to select the first restraint you want to visualize in the table
- Scroll up and down the table with the arrow keys to search for restraints
- NEW in MakeTNT2.1.1: Click on an atom in the Jmol-window to highlight the restraints it is involved in
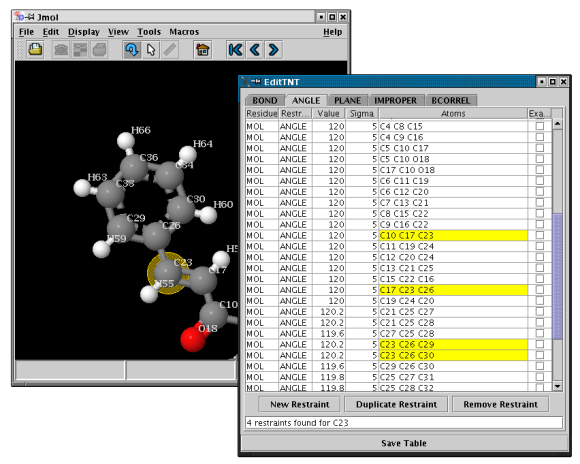
- Double click on a cell in order to edit restraints
- The display will update as soon as you press ENTER or move to
another cell
- For planarity restraints atom counts will automatically update upon the modification of atom records
- Check the display before you save the table
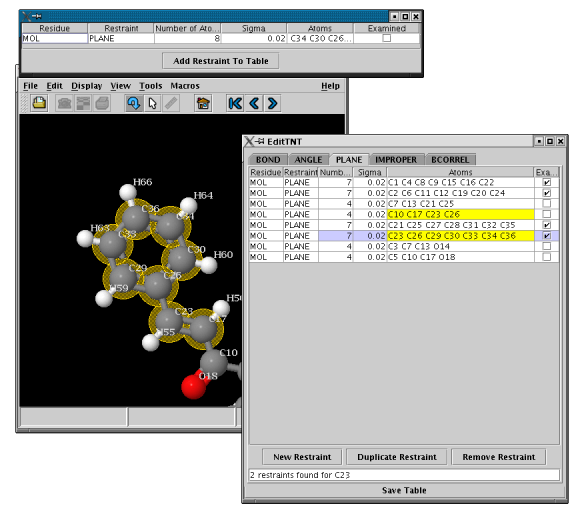
- NEW in MakeTNT2.1.1: Interactive Addition/Duplication of Restraints
- Click on the "Add Restraint" or "Duplicate Restraint" - button in the main window
- Click on atoms in the Jmol-display to select atoms for the new restraint, they will show up
in the extra table
- Click on a selected atom (NO DOUBLE CLICK) to remove it again
- Insert the new restraint into the main table by clicking on "Add Restraint To Table"
- Gelly optimization:
- Click on "Run Gelly" to optimize your structure to the restraints chosen (only possible if
pdb-file was submitted)
- Once the optimization is complete (< 10s), switching to the optimized conformation (and back) will
become available
- Optimization will always start from the original pdb-file
- If you would like an optimization to start from a previous gelly run, please restart
EditTNT with the gelly-output PDB-file and the modified dictionary (*.gelly)
- Please also notice the messages provided by EditTNT
- To use gelly for changing chiralities, check the example below
If you click on Save Table on the menu, the table will be saved
into a file with the extension ".user". If you have chosen to optimize a structure with Gelly, the latest update of the table will be saved into a file with the extension ".gelly", the optimized structure will be saved into a file with the extension ".opt.pdb" and a file "gelly_refine.log" will show you the course of the gelly - optimization and its final agreement with the restraints chosen by you.
Please copy the folder samples/maketnt of your MakeTNT-toolkit distribution over:
|
% cp -pri $BDG_home/MakeTNT/samples/maketnt .
|
This will create a local copy of the examples that we provide. In this
folder you will find restraint files generated by Corina,
the Libcheck
program (part of Refmac
and CCP4) and xplo2d
(XPLOR option), as well as the corresponding TNT-dictionary files
converted by MakeTNT. For details of MakeTNT usage, please refer to the MakeTNT-documentation.
To look at TNT-restraints generated for the compound taxotere by
Corina and converted into TNT-format by MakeTNT, please go to the
maketnt/CORINA directory and type:
|
% EditTNT taxotere.dic TXL.mol
|
This will start the interface as shown above. Please select
PLANE in the tab-header section. Click on an atom
to highlight the planar restraints it is involved in. A count
of restraints found for this atom will appear in the text window
towards the bottom of EditTNT.
You can also use the mouse keys to scroll up and down the list of plnar
restraints.
Looking at the table you will find, that for this molecule all PLANE
restraints are computed in a very satisfactory way, as is usually the
case when using Corina-derived parameters. In the next sections, we
will learn how to edit and - if necessary - modify restraints
generated by other sources.
To view TNT-restraints derived from a Libcheck/Refmac descriptions,
please go to the maketnt/Refmac directory and type:
|
% EditTNT adenosinetriphosphate.dic ATP.pdb
|
Select the PLANE-table again and look at the only entry,
which describes the planarity of the ATP-purine ring. If you wish to
add the N6 atom to this list, double click on the cell with the Header
Atoms. It will change colour and you can scroll to either end
of the list using the arrow keys on your keyboard. You can delete text
in the cell using the backspace key and add text by just typing it
into the cell. To add the "N6" atom to the list, go to the end of it
and type ", N6".
Alternatively, click on "Duplicate Restraint" to start a new table with
the same atom atoms. To change the selection of atoms, either
- click on the atom in the JMOL display window, this will
- selected atoms
- deselect selected atoms
or
- double click in the atoms cell to edit, a single click outside this cell,
but inside the table will re-enable selection from the molecular viewer
Once you are happy with your new restraint, insert it into the main table by
pressing the respective button and remove the original restraint from there.
A similar mechanism can be used to add new restraints using the "Add Restraint" button.
To save the new restraint list, select Save Table.
The TNT-dictionary file will be saved using the original name with the
extension ".user". Before saving the list, please press "Enter" to
make sure that all the latest changes are included.
For a demonstration of TNT-restraints derived from xplo2d
(XPLOR-format), please go to the maketnt/XPLOR directory and
type:
|
% EditTNT rosiglitazone.dic BRL.pdb
|
Please select the PLANE-table again. You will see four
different PLANE-restraints generated for rosiglitazone. The first
restraint describes the planar thiazolidinedione group and is correct,
as are restraints 2 and 4 (for the benzyl and pyrdinyl group,
respectively). However, the third restraint (4 atoms) is derived from
a particular conformation rather than from a chemical rule. If you are
interested in refining rosiglitazone in conformations different to the
one in the structure shown, you probably want to remove that
restraint. To do so, simply select it and click on the Remove
Restraint - button. This removes the restraint in the table, but
not in the underlying file. To make the change permanent, please save
the new restraints by clicking on Save Table.
To find out how to use gelly interactively within EditTNT, please
enter the maketnt/PDB2TNT directory and
type:
|
% EditTNT TXL.dic TXL_tnt.pdb
|
Now, let's assume you would like to invert the chirality around the C34 atom.
To do that, you enter the IMPROPER card in
EditTNT, select the 8th restraint from the top, change its value
from -30.7 to 30.7, press the ENTER key on the keyboard and then
press the "Run Gelly" button on the interface. You will get a message
that gelly has started running.
Once optimization is complete (<10s), you will
be notified by an EditTNT-message. Simultanously, a button for loading the
new structure into EditTNT/JMOL will become active. Another button will
take you back to the original structure.
The "Run-Gelly" button could be modified by an option to specify parts of the
molecule to be fixed during the optimization. Please let us know whether
such an option would be useful to you.
Maria Brandl,
<buster-develop@GlobalPhasing.com>
Last modification: 10.07.08

