Further rhofit tests.
Content:
- introduction and methods
- rhofit results
- summary
1. methods
A range of structures were selected from the pdb encompassing a range of resolution, ligand size and complexity.
Rather than directly using the 2Fo-Fc density map from the refined model the following procedure was adopted:
- The relevant ligand was striped out from the pdb entry.
- The protein was then refined by
refine -p molrep.pdb -m data.mtz -autoncs -d FIRST
FitMAP -p FIRST/refine.pdb -m FIRST/2Fo-Fc.map -dm FIRST/Fo-Fc.map
refine -p protein_fitted.pdb -m data.mtz -autoncs -L -d SECOND
This results in water insertion being applied during refinement. Before the final cycle likely areas for ligand binding are found and the model water molecules in this area are removed from the model, but kept for the purposes of defining the bulk solvent mask. This is found to improve difference density.
- Model ligand coordinates were obtained from the PDBeChem service. This generates model coordinates using the CORINA program.
- The model ligand was then placed into density using rhofit. Unless stated otherwise rhofit was run with the command:
rhofit -lp ideal.pdb -m refine.mtz -p refine.pdb -d RHOFIT
2. rhofit results
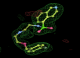
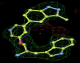
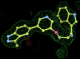
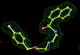
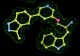
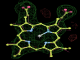
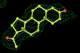
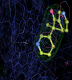
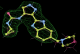
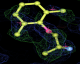
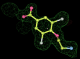
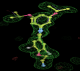
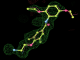
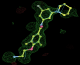
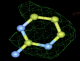
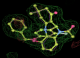
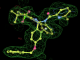
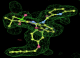
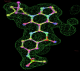
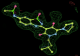
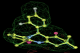
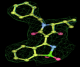
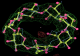
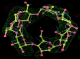
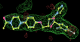
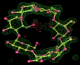
- Click on the picture to expand (or right click to get in another tab)! Difference density as found by autoBUSTER refine -L contoured at 3.0 sigma in green, rhofit best solution in atom-colored ball and stick. pdb ligand position in blue/purple lines.
- Both heme and ligand removed prior to refinement. rhofit run initially with heme (with -clashweight 0 to turn off protein clash detection). After correct heme solution placed in the protein, rhofit then run a second time with the ligand.
- Initial run failed due to no density for terminal cyclohexyl ring in ligand - failure as volume of cluster < 80* volume of ligand. Ran
- prep_rhofit -p refine.pdb -m refine.mtz -V <vol>
- where <vol> was ~70% ligand volume. Re-ran rhofit as
- rhofit -lp ligand.pdb -m refine.mtz -protein refine.pdb -c cluster_1.pdb
- Clustering/water addition in autobuster was not entirely successful in this case (as the map illustrates), due to the map. Nevertheless, rhofit has correctly placed the ligand!
- alternate conformations built in deposited structure.
- exact pdb solution not quite found - due to density (from an ordered water) at lower end of the ligand.
- Initially, expected solution not quite found due to dictionary issue. On correcting dictionary and re-running rhofit with the -l dictionary option, the correct solution was built.
- See (2). Both BCD and KLN removed. rhofit run first with BCD in 2 clusters. After building BCD into protein, rhofit re-run with KLN in 2 clusters.
3. Summary
Out of 24 tests, the correct answer was found in ALL cases and as the top solution in all but 3 cases.
Page by Andrew Sharff 11 November 2009. Address problems, corrections and clarifications to buster-develop@globalphasing.com