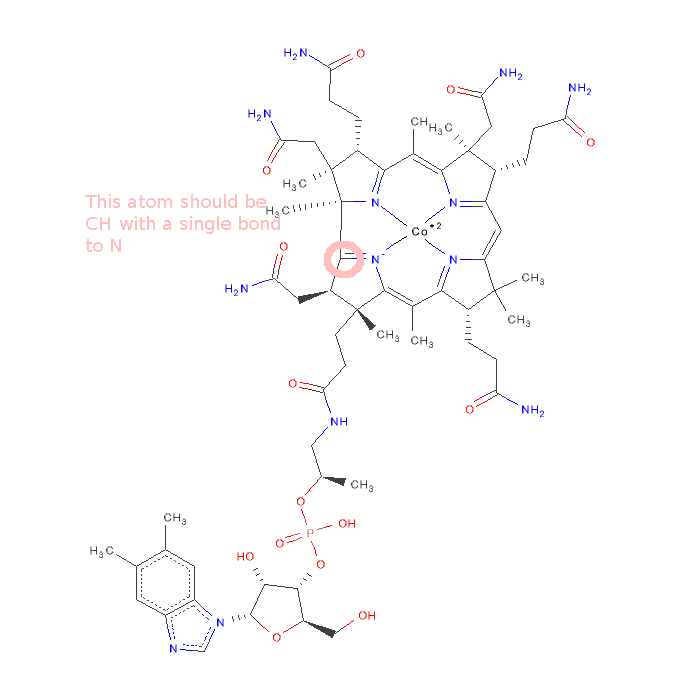
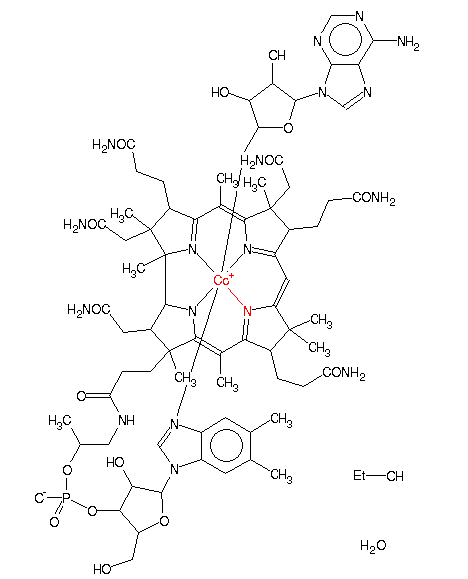
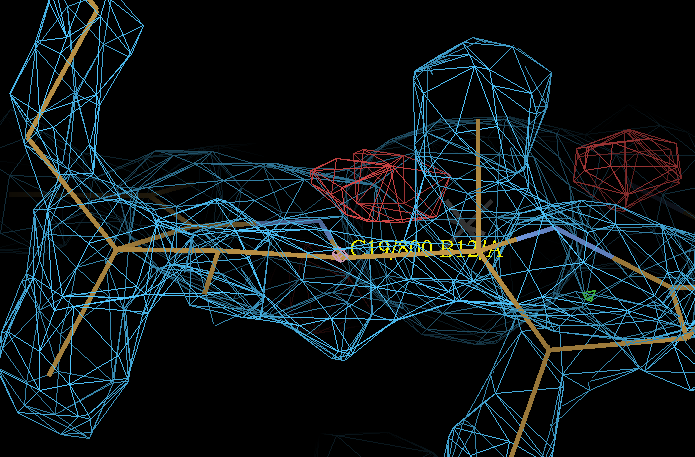
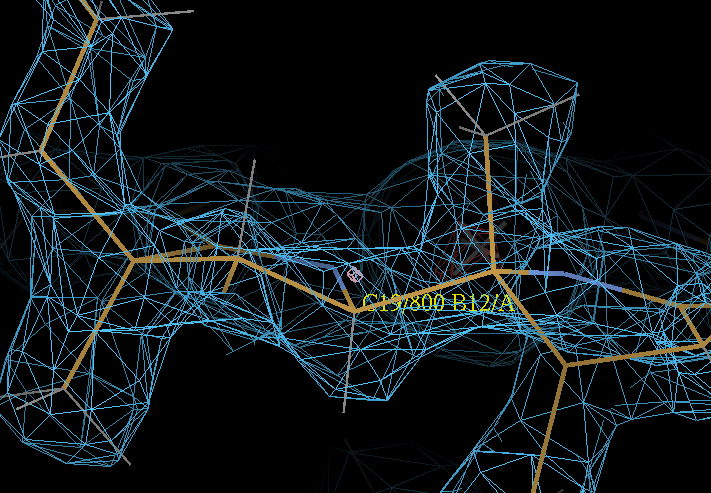
| Attachments | |
|---|---|
| PAFBUV.png | 7K |
| 3ci1_00_maponly-C19.png | 170K |
| 3ci1_02_plush-C19.png | 180K |
| B12_check.tsv | 4K |
| 3ci1_02_plush.pdb | 291K |
| 3ci1_00_maponly.mtz | 1MB |
| PAFBUV_pdbatomnames_inc5AD.pdb | 16K |
| PAFBUV.pdb | 23K |
| B12.cif | 40K |
| B12_D3L0.gif | 8K |
| 5AD.grade_PDB_ligand.cif | 10K |
| 3ci1.pdb | 190K |
| 3ci1_02_plush.mtz | 1MB |