
| Attachments | |
|---|---|
| 3nrz.wrongstart.png | 8K |
| 3nrz.rightpostref.png | 8K |
| insulin-five.png | 8K |
| insulin-six.png | 8K |
| 3nrz.wrongpostref.png | 9K |
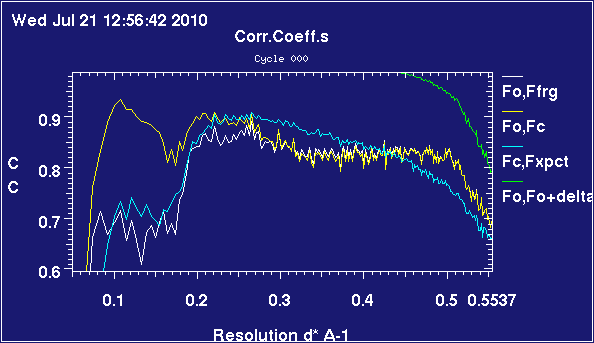
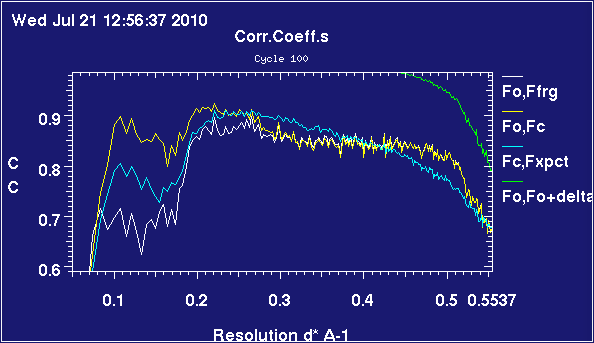
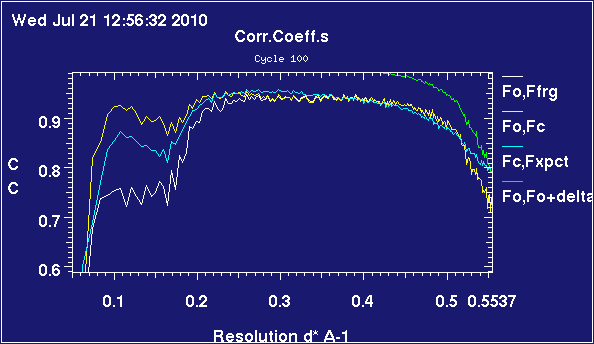
These plots are obtained from 2omg, which is a trimer of insulin heterodimers. It's been refined twice, once complete and once with chain C deleted.
The relevant difference is that, for the map with a substantial missing part, the refinement is having difficulty matching data in the lower resolution bins - observe that, whilst the correlations are above 90% almost all the way in the case with six chains present, they are below 80% up to 0.2 inverse-angstrom when one chain is missing. The correlations are very similar above 0.4 inverse-angstrom, corresponding to details on the order of scale of side-chains.
In this example, the input model is missing about sixty residues, both at termini of several chains and in internal loops, which show up very well-ordered in the electron density.
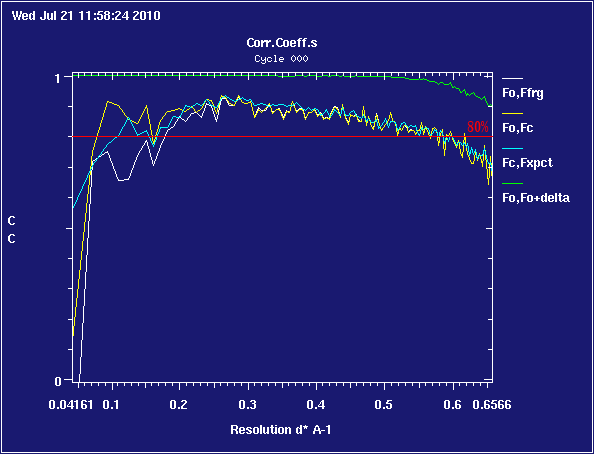
 The correlation left of 0.2 A-1 is a bit improved, but note that the blue line still crosses the yellow one
The correlation left of 0.2 A-1 is a bit improved, but note that the blue line still crosses the yellow one
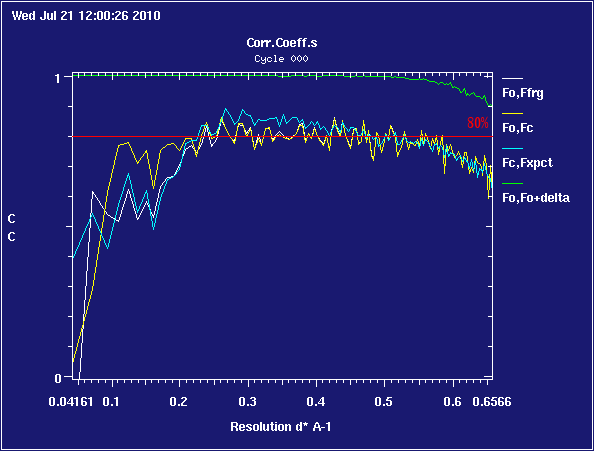
 Notice that the blue line now tracks the yellow one much more closely
Notice that the blue line now tracks the yellow one much more closely