Content:
Diffraction limits & principal axes of ellipsoid fitted to diffraction cut-off surface:
1.955 0.9571 0.0000 0.2898 a* + 0.020 c*
1.940 0.0000 1.0000 0.0000 b*
1.906 -0.2898 0.0000 0.9571 -0.505 a* + 0.863 c*
| Date deposited | Date data collection | Resolution | R, Rfree |
| 20110821 | 20100806 | 1.99 | 0.1920 0.2350 |
Molprobity (CCP4 7.0 version) summary:
Ramachandran outliers = 0.00 %
favored = 98.36 %
Rotamer outliers = 1.52 %
C-beta deviations = 1
Clashscore = 4.19
RMS(bonds) = 0.0156
RMS(angles) = 1.40
MolProbity score = 1.34
Resolution = 1.99
R-work = 0.1920
R-free = 0.2350
Additional analysis:
Number of waters = 266 <B> (all atoms) = 27.84 ( sd = 9.19 ) for 2677 non-hydrogen atoms <B> (protein) = 27.55 ( sd = 9.00 ) for 2377 non-hydrogen atoms <B> (water) = 29.32 ( sd = 10.10 ) for 266 non-hydrogen atoms <B> (others) = 36.95 ( sd = 8.87 ) for 34 non-hydrogen atoms B min/max (all non-hydrogen atoms) = 10.41 / 69.61 B min/max (protein non-hydrogen atoms) = 11.90 / 69.61 B min/max (water non-hydrogen atoms) = 10.41 / 57.93 B min/max (other non-hydrogen atoms) = 20.48 / 46.19
Molprobity (CCP4 7.0 version) summary:
Ramachandran outliers = 0.00 %
favored = 97.70 %
Rotamer outliers = 1.14 %
C-beta deviations = 0
Clashscore = 2.09
RMS(bonds) = 0.0117
RMS(angles) = 1.53
MolProbity score = 1.09
Resolution = 1.99
R-work = 0.1847
R-free = 0.2208
Additional analysis:
Number of waters = 326 <B> (all atoms) = 30.18 ( sd = 10.73 ) for 2737 non-hydrogen atoms <B> (protein) = 28.40 ( sd = 9.32 ) for 2377 non-hydrogen atoms <B> (water) = 42.24 ( sd = 12.38 ) for 326 non-hydrogen atoms <B> (others) = 39.20 ( sd = 5.74 ) for 34 non-hydrogen atoms B min/max (all non-hydrogen atoms) = 13.19 / 80.90 B min/max (protein non-hydrogen atoms) = 14.27 / 77.13 B min/max (water non-hydrogen atoms) = 13.19 / 80.90 B min/max (other non-hydrogen atoms) = 29.09 / 45.52
Refinement progression:
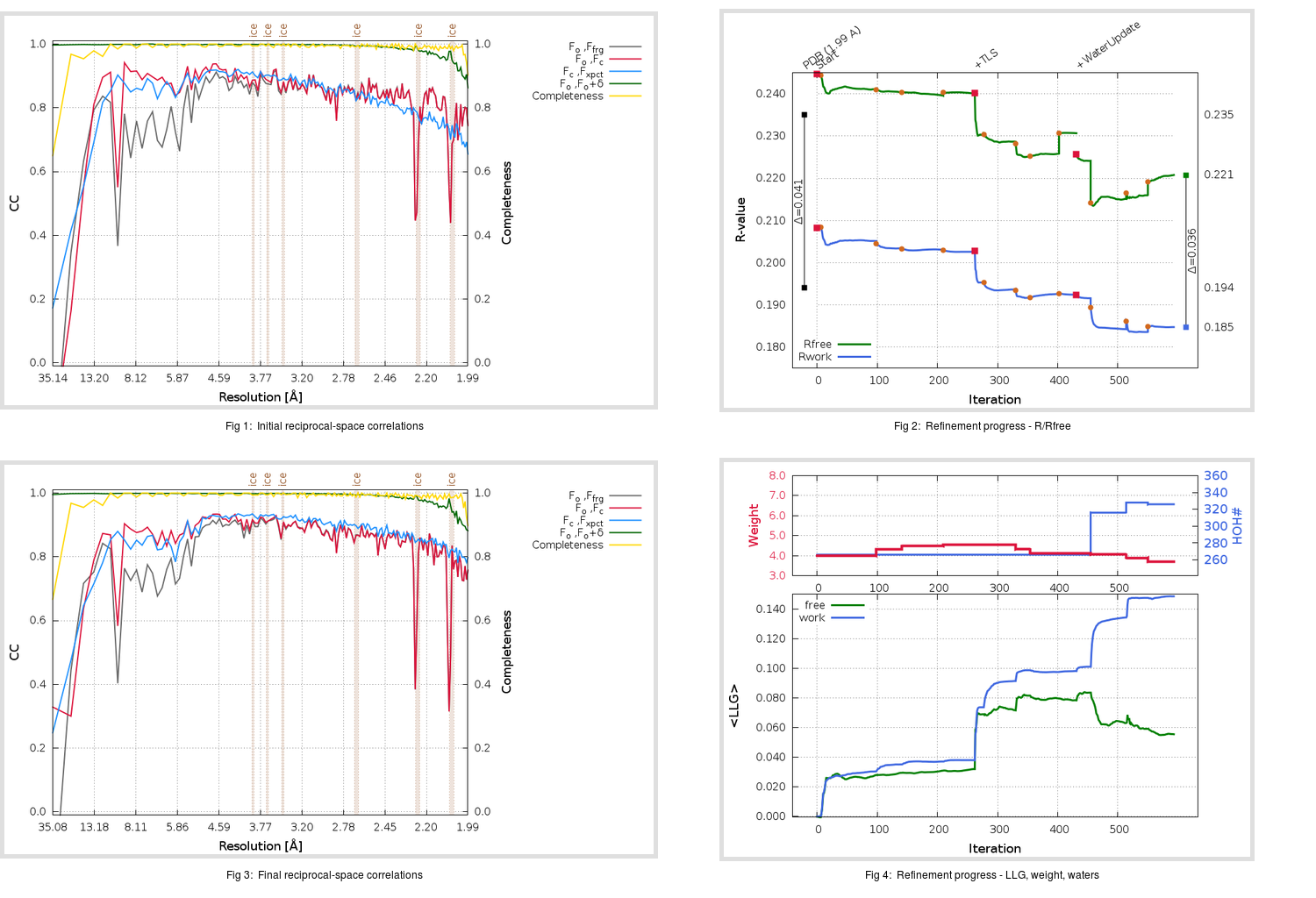 |
Results:
| File | Remark |
| 3TIT_aB_refine.01_03_refine.pdb.gz | exact refinement commands are in header |
| 3TIT_aB_refine.01_03_refine.mtz.gz | including original deposited data and several re-refinement map coefficients |
| 3TIT_aB_refine.01_03_BUSTER_model.cif.gz | including any non-standard compound restraints |
| 3TIT_aB_refine.01_03_BUSTER_refln.cif.gz |