Content:
Diffraction limits & principal axes of ellipsoid fitted to diffraction cut-off surface:
2.514 0.7149 0.0000 -0.6992 0.571 a* - 0.821 c*
2.951 0.0000 1.0000 0.0000 b*
2.935 0.6992 0.0000 0.7149 0.596 a* + 0.803 c*
| Date deposited | Date data collection | Resolution | R, Rfree |
| 20200218 | 20200211 | 2.68 | 0.1970 0.2290 |
Molprobity (CCP4 7.0 version) summary:
Ramachandran outliers = 0.00 %
favored = 95.53 %
Rotamer outliers = 3.97 %
C-beta deviations = 0
Clashscore = 5.54
RMS(bonds) = 0.0041
RMS(angles) = 0.99
MolProbity score = 2.07
Resolution = 2.68
R-work = 0.1970
R-free = 0.2290
Additional analysis:
Number of waters = 26 <B> (all atoms) = 108.85 ( sd = 28.04 ) for 13180 non-hydrogen atoms <B> (protein) = 108.05 ( sd = 27.77 ) for 12782 non-hydrogen atoms <B> (water) = 82.32 ( sd = 14.01 ) for 26 non-hydrogen atoms <B> (others) = 138.01 ( sd = 20.66 ) for 372 non-hydrogen atoms B min/max (all non-hydrogen atoms) = 52.15 / 250.89 B min/max (protein non-hydrogen atoms) = 52.15 / 250.89 B min/max (water non-hydrogen atoms) = 60.44 / 116.90 B min/max (other non-hydrogen atoms) = 75.04 / 196.78
Molprobity (CCP4 7.0 version) summary:
Ramachandran outliers = 0.13 %
favored = 96.43 %
Rotamer outliers = 7.36 %
C-beta deviations = 9
Clashscore = 3.94
RMS(bonds) = 0.0127
RMS(angles) = 1.58
MolProbity score = 2.08
Resolution = 2.68
R-work = 0.1927
R-free = 0.2184
Additional analysis:
Number of waters = 196 <B> (all atoms) = 110.00 ( sd = 25.47 ) for 13350 non-hydrogen atoms <B> (protein) = 109.14 ( sd = 24.57 ) for 12782 non-hydrogen atoms <B> (water) = 91.83 ( sd = 15.68 ) for 196 non-hydrogen atoms <B> (others) = 149.27 ( sd = 25.85 ) for 372 non-hydrogen atoms B min/max (all non-hydrogen atoms) = 63.36 / 215.18 B min/max (protein non-hydrogen atoms) = 65.99 / 211.66 B min/max (water non-hydrogen atoms) = 63.36 / 215.18 B min/max (other non-hydrogen atoms) = 81.10 / 190.98
Refinement progression:
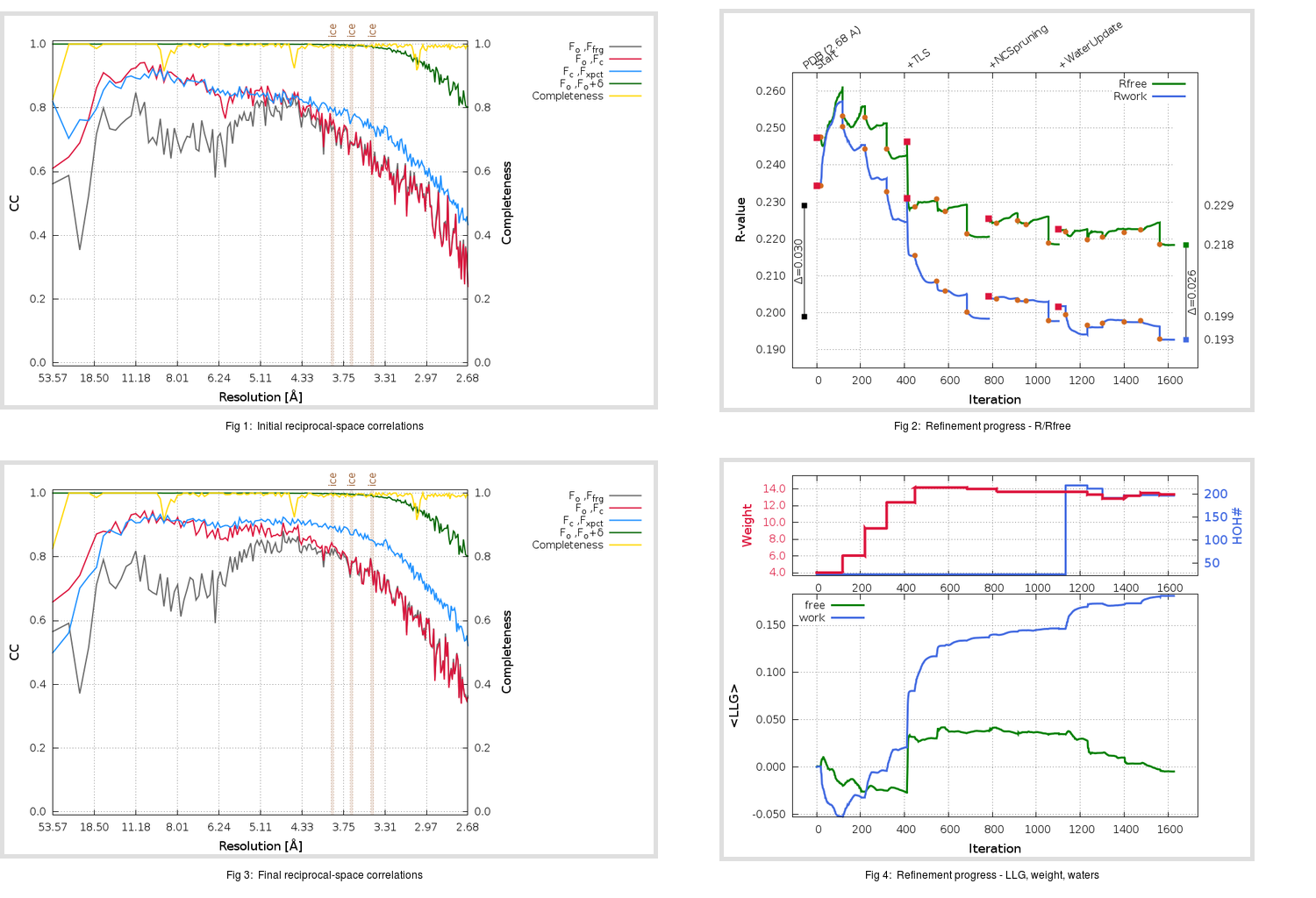 |
Results:
| File | Remark |
| 6VW1_aB_refine.01_04_refine.pdb.gz | exact refinement commands are in header |
| 6VW1_aB_refine.01_04_refine.mtz.gz | including original deposited data and several re-refinement map coefficients |
| 6VW1_aB_refine.01_04_BUSTER_model.cif.gz | including any non-standard compound restraints |
| 6VW1_aB_refine.01_04_BUSTER_refln.cif.gz |