
WORK-IN-PROGRESS
Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2. Kim, Y., Jedrzejczak, R., Maltseva, N.I., Endres, M., Godzik, A., Michalska, K., Joachimiak, A.
Data was collected on 19th February 2020 on beamline 19-ID (SBC-CAT, APS) on a Pilatus3 6M and refined to 2.20 A using Phenix.
We have 200 images of 0.5 deg/image (i.e. 100 degrees of data) with 0.4 sec/image. Following the theme of high-dose, low-multiplicity, wide-slicing, these parameters look again to be the exact opposite from current recommendation for collecting good data on these detectors.
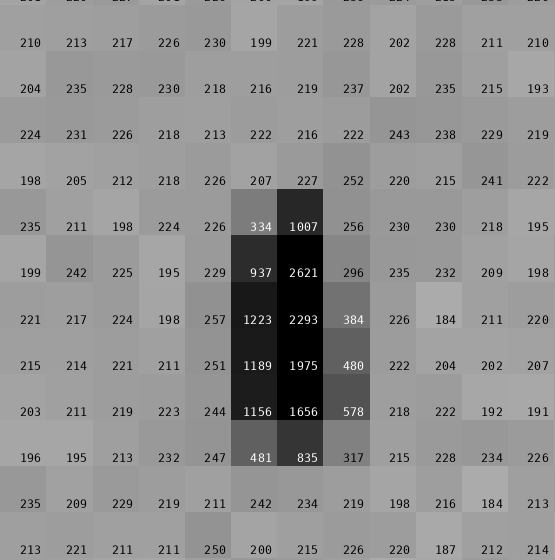
Running
process ReverseRotationAxis=yesgives us
Spacegroup name P63
Unit cell parameters 150.375 150.375 111.102 90.0 90.0 120.0
Wavelength 0.97918 A
Diffraction limits & principal axes of ellipsoid fitted to diffraction cut-off surface:
1.985 1.0000 0.0000 0.0000 0.894 _a_* - 0.447 _b_*
1.985 0.0000 1.0000 0.0000 _b_*
2.158 0.0000 0.0000 1.0000 _c_*
Criteria used in determination of diffraction limits:
-----------------------------------------------------
local(I/sigI) >= 1.20
Overall InnerShell OuterShell
---------------------------------------------------------------------------
Low resolution limit 45.003 45.003 2.091
High resolution limit 1.985 5.687 1.985
Rmerge (all I+ & I-) 0.127 0.045 1.136
Rmeas (all I+ & I-) 0.140 0.049 1.275
Rpim (all I+ & I-) 0.059 0.021 0.571
Total number of observations 480926 23831 20481
Total number unique 85630 4283 4283
Mean(I)/sd(I) 9.9 26.8 1.7
Completeness (spherical) 87.0 99.8 30.2
Completeness (ellipsoidal) 93.9 99.8 54.0
Multiplicity 5.6 5.6 4.8
CC(1/2) 0.997 0.999 0.328
 |
 |
 |
We have high enough completeness to also check for radiation damage later in BUSTER:
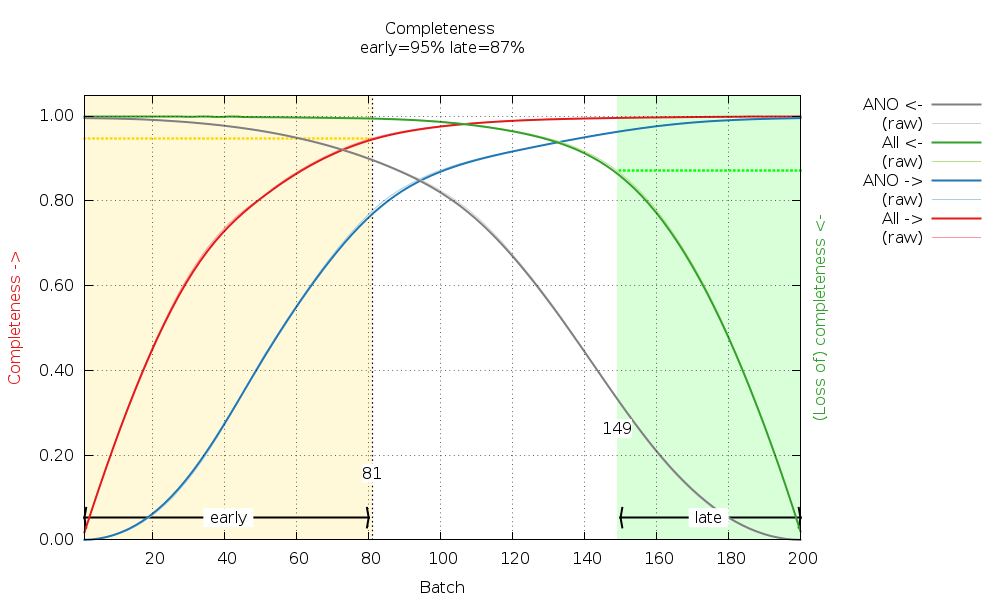
There are some warnings about possible twinning (from POINTLESS): Some tests suggest that the data might be twinned (alpha = 0.156-0.175).
The 1.9 A Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with a Citrate Kim, Y., Jedrzejczak, R., Maltseva, N., Endres, M., Godzik, A., Michalska, K., Joachimiak, A., Center for Structural Genomics of Infectious Diseases (CSGID) To be published.
Data was collected on 25th February 2020 on beamline 19-ID (SBC-CAT, APS) on a Pilatus3 6M and refined to 1.90 A using Phenix.
We have two sweeps of data:
Nsp15_M4_G4_RNA_eg_data_#####.cbf : 1 - 52 Nsp15_M4_G4_RNA_eg_data2_#####.cbf : 1 - 130
with a 65 second gap between them - so probably a change of position on the same crystal? Although, the final image 52 is actually empty, so maybe some other hic-up and the second sweep is just a continuation. Anyway, the second sweep starts at the Omega angle of image 51 (of the first sweep) - giving us 90 degrees of data.
Both were collected as again as high-dose (bacground pixels with counts of about 60), low-multiplicity (90 degrees of data), wide-slicing (0.5 deg/image)data ... unfortunately.
We can process the data as it is - meaning we would
An alternative is to bring this together into a single scan via some symbolic links:
mkdir Images2 cd Images2 n=0 for i in `seq -w 1 50` do n=`expr $n + 1` ln -s ../Images/Nsp15_M4_G4_RNA_eg_data_000$i.cbf . done for i in `seq -w 1 130` do n=`expr $n + 1` ii=`echo $n | awk '{printf("%3.3d",$1)}'` ln -s ../Images/Nsp15_M4_G4_RNA_eg_data2_00$i.cbf Nsp15_M4_G4_RNA_eg_data_00$ii.cbf done
And then process with
process -I Images2 ReverseRotationAxis=yesgives us
Spacegroup name P63
Unit cell parameters 150.655 150.655 111.555 90.0 90.0 120.0
Wavelength 0.97918 A
Diffraction limits & principal axes of ellipsoid fitted to diffraction cut-off surface:
1.752 1.0000 0.0000 0.0000 0.894 _a_* - 0.447 _b_*
1.752 0.0000 1.0000 0.0000 _b_*
2.104 0.0000 0.0000 1.0000 _c_*
Criteria used in determination of diffraction limits:
-----------------------------------------------------
local(I/sigI) >= 1.20
Overall InnerShell OuterShell
---------------------------------------------------------------------------
Low resolution limit 45.103 45.103 1.859
High resolution limit 1.752 5.098 1.752
Rmerge (all I+ & I-) 0.073 0.036 0.579
Rmeas (all I+ & I-) 0.081 0.040 0.707
Rpim (all I+ & I-) 0.034 0.017 0.398
Total number of observations 557214 29665 17009
Total number unique 114261 5714 5714
Mean(I)/sd(I) 12.4 34.3 1.8
Completeness (spherical) 79.4 95.6 24.5
Completeness (ellipsoidal) 94.8 95.6 67.5
Multiplicity 4.9 5.2 3.0
CC(1/2) 0.998 0.999 0.651
and
 |
 |
 |
So incomplete data (low-multiplicity) can achieve quite nice pictures (see the one to the right above), but that is not necessarily the primary goal here ;-)
Files: