
Content
These pages are intended as a resource for the tutorials of our software at the International workshop, Joint CeBEM-CCP4 initiative, April 9th-17th, 2013, Institut Pasteur Montevideo, Uruguay .
Much more introductory material is available for each program:
autoPROC relies on external programs like:
All programs should be configured and setup correctly. You can test this by running
% process -h
For running this tutorial, it might be a good idea to start in a fresh, empty directory.
This should be a reasonably fast run through all stages of data-processing (about 5 min on an i7-2720QM laptop with 8Gb of memory).
We're going to use the data for 1O22 from the JCSG. It is
Note: more examples and tutorials are given on the autoPROC wiki. And make sure you have a copy of the autoPROC reference card.
This peak dataset consists of 90 images (tm0875_8p44_1_E1_001.img to tm0875_8p44_1_E1_090.img). A quick look at the image header can be done with
% imginfo ~/Desktop/Tutorials/1o22/tm0875_8p44_1_E1_001.img
that gives
>>> Image format detected as ADSC ===== Header information: date = 13 Oct 2002 18:47:43 exposure time [seconds] = 45.000 distance [mm] = 200.000 wavelength [A] = 0.977800 Phi-angle (start, end) [degree] = 90.000 91.000 Oscillation-angle in Phi [degree] = 1.000 Omega-angle [degree] = 0.000 2-Theta angle [degree] = 0.000 Pixel size in X [mm] = 0.102400 Pixel size in Y [mm] = 0.102400 Number of pixels in X = 2048 Number of pixels in Y = 2048 Beam centre in X [mm] = 104.900 Beam centre in X [pixel] = 1024.414 Beam centre in Y [mm] = 104.800 Beam centre in Y [pixel] = 1023.438 Overload value = 65535
It is a good idea to have two terminals open: one for running the program and a second to look at results already while its running (in case you are impatient).
We can run this with all defaults
% process -I ~/Desktop/Tutorials/1o22 -d 00 | tee 00.lis
or with some additional automation using the process command from a terminal/shell:
% process -M automatic -I ~/Desktop/Tutorials/1o22 -d 01 | tee 01.lis
which should give us after about 3 minutes a processed dataset (using XDS, POINTLESS and SCALA/AIMLESS as part of autoPROC). There are probably a few interesting notes along the way ... and in your second terminal you could already have a look at intermediate results, eg. with
% cd 00 % ls -ltra % ls -ltra *.png % display distance.png ...
Since the specification of the beam centre (or detector) is a crucial part, some information is given:
 |
It seems there is some problem regarding indexing - autoPROC enters the so-called "iterative indexing" mode:
 |
Remember, we would always like to index all spots on the images - anything not indexed is something to look at:
 |
The above rough resolution limits are based on spots - where a spot is most often much stronger than a still useful reflection: so the high-resolution limit is probably conservative.
By default, the initial indexing is done in P1. Hopefully we reach a good indexing solution with small errors:
 |
The first list of possible indexing solutions for different lattices already give an indication of the spacegroup:
 |
After the initial integration (in P1) we use POINTLESS to decide on most likely spacegroup:
 |
It is always a good idea to check the above analysis very carefully: are all symmetries defined with the same probability? Was there enough data available to make such a crucial decision about spacegroup?
A quick analysis of data quality using the chosen spacegroup can eg already give an indication of anomalous signal:
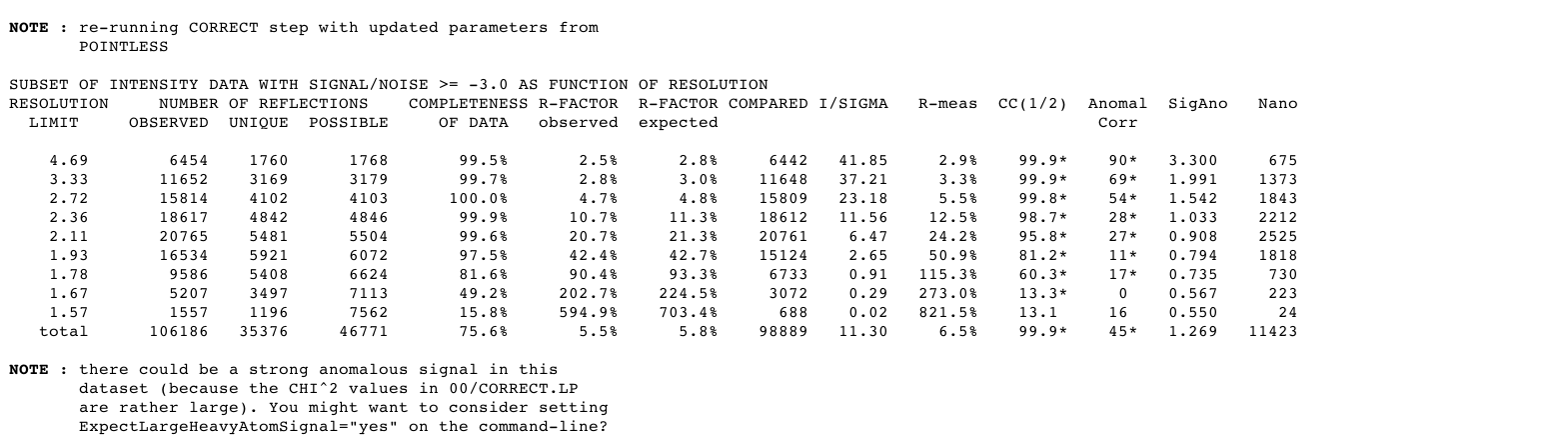 |
Another round of integration is done, now in the higher symmetry spacegroup:
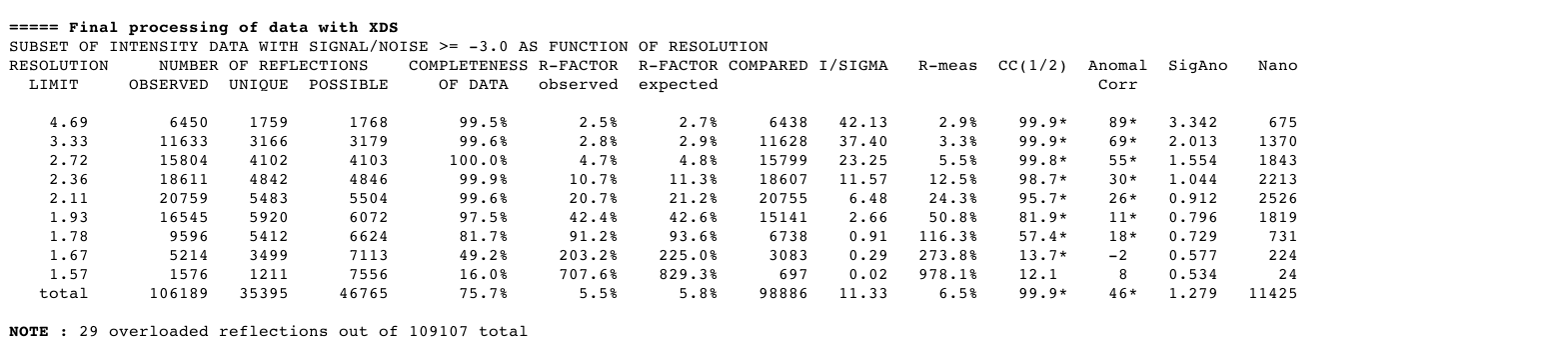 |
We follow this with a scaling cycle using AIMLESS:
 |
resulting in a "table 1" set of statistics:
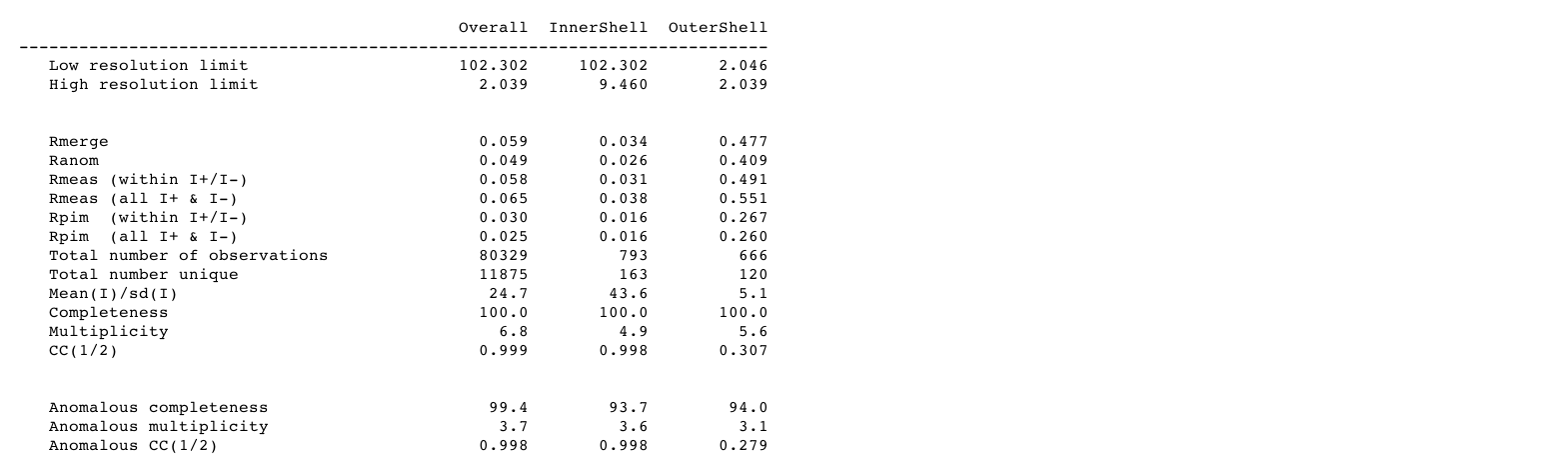 |
In case of severe anisotropy, AIMLESS will give some indication of the problem:
 |
Finally, we get a processed reflection file and the required section for structure deposition (REMARK 200):
 |
You should also have a look at the various graphs (PNG files) eg. with
% display 00/*.png
What do they show? Anything interesting in those images? What shape would we expect and where do we get deviations from that expectaions?
You should also look at the scaling logfile using:
% loggraph 00/aimless.log
Make sure you know where to find documentation and help online:
% process -h % refine -h
Ideally just running e.g.
% process -d 01 | tee 01.lis
in your directory with the images should be enough. However, often it is necessary to provide some additional, non-default parameters to describe the beamline/instrument - see here for some more information.
If you have potentially difficult data, you could also try and run with
% process -M LowResOrTricky -d 02 | tee 02.lis
Please see the autoPROC reference card (on the course moodle site) and the autoPROC wiki for more help - or type
% process -h % process -M list
Original page: April 2013, Clemens Vonrhein