Charging¶
(de)protonating groups likely to be charged at neutral pH¶
Molecular Databases like the wwPDB Chemical Component Dictionary normally have neutral forms of compounds that can be expected to be charged at neutral pH. For instance, carboxylic acids are protonated rather than forming a carboxylate ion.
By default, Grade2 examine the input molecule and charge a number
of common groups by adding or removing a proton. If a group
is charged then a WARNING message will be written to the terminal
output (please see
WARNING about charging example output).
If the charging is not wanted then use the --no_charging
command-line option and no alteration will be made to the input molecule.
Currently, Grade2 will charge the groups listed in the sections below.
If you want to model a ligand with a protonation state that is distinct from the standard charging scheme then use manual editing with Mercury as demonstrated by the FAQ How can I produce restraints for a ligand with a different protonation state or tautomer?.
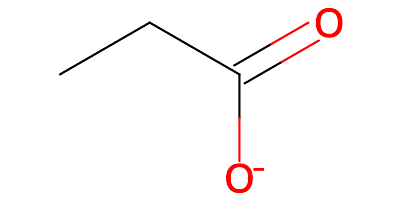
Carboxylic acid(s) to carboxylate ion(s)¶
A SMARTS pattern [$([OX2H1][CX3]=O)] is used to detect neutral carboxylic
acid and the proton is removed to leave a carboxylate ion.
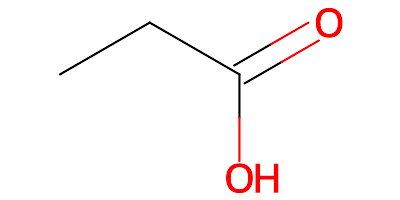
For example, running grade2 for the PDB component
PPI https://www.rcsb.org/ligand/PPI propanoic acid:
$ grade2 --PDB_ligand PPI
Produces the warning messages:
WARNING: Charging groups likely to be charged at neutral pH.
WARNING: ---- If you do not want this, rerun with the option: -N, --no_charging
charging carboxylic acid(s) to carboxylate ion(s)
remove proton HO2 and add negative charge to atom O2
The final molecule will be the carboxylate:
rather than the original neutral PPI component:
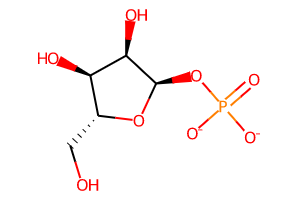
Phosphoric acid(s) to phosphate ion(s)¶
A SMARTS pattern [$([OX2H1][PX4]=O)] is used to detect phosphoric
acids with a hydrogen atom and the proton is removed to leave a phosphate ion.
If a phosphoric acid has multiple hydrogen atoms attached all are removed.
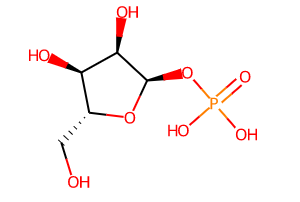
For example, running grade2 for the PDB component
R1P https://www.rcsb.org/ligand/R1P ribose-1-phosphate:
$ grade2 --PDB_ligand R1P
Produces the warning messages:
WARNING: Charging groups likely to be charged at neutral pH.
WARNING: ---- If you do not want this, rerun with the option: -N, --no_charging
charging phosphoric acid(s) to phosphate ion(s)
remove proton HOP2 and add negative charge to atom O2P
remove proton HOP3 and add negative charge to atom O3P
The final molecule will be the ribose-1-phosphate dianion with both the protons removed:
rather than the original neutral R1P component:
Alkylamine to alkylammonium¶
A SMARTS pattern [$([NX3;H2][CX4])] is used to detect nitrogen atoms
with 2 hydrogen atoms attached that are also bonded to an SP3 carbon atom.
A proton hydrogen atom is added in these cases.
For example, running grade2 for the PDB component
01R https://www.rcsb.org/ligand/01R:
$ grade2 --PDB_ligand 01R
Produces the warning messages:
WARNING: Charging groups likely to be charged at neutral pH.
WARNING: ---- If you do not want this, rerun with the option: -N, --no_charging
charging alkylamine to alkylammonium
add new proton H3 onto atom N19 (existing hydrogen atom_ids: H19, H2)
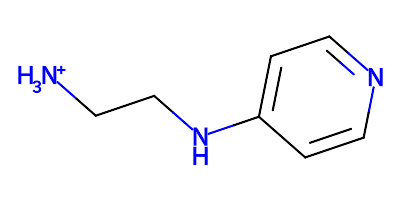
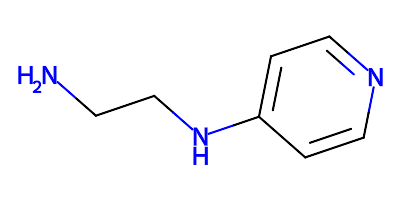
Notice that the name of the proton hydrogen added is given. The result is a cation:
rather than the original neutral 01R component:
notice that only the terminal -CH2-NH2 nitrogen atom is protonated with
the other nitrogen atoms left alone.
Dialkylamine to dialkylammonium¶
A SMARTS pattern [$([NX3;H1]([CX4])[CX4])] is used to detect nitrogen atoms
with a hydrogen atom attached that are also bonded to two SP3 carbon atoms.
A proton hydrogen atom is added in these cases.
For example, running grade2 for the PDB component
PIP https://www.rcsb.org/ligand/PIP piperidine:
$ grade2 --PDB_ligand PIP
Produces the warning messages:
WARNING: Charging groups likely to be charged at neutral pH.
WARNING: ---- If you do not want this, rerun with the option: -N, --no_charging
charging dialkylamine to dialkylammonium
add new proton HN1B onto atom N1 (existing hydrogen atom_ids: HN1)
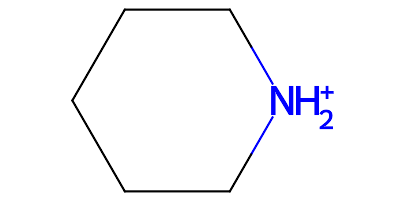
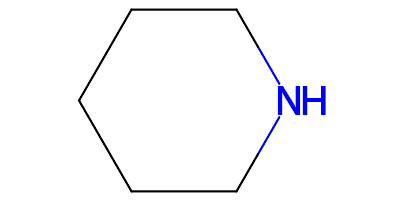
Notice that the atom name of the proton hydrogen added is based on the atom name of the existing hydrogen. The resulting molecule is a cation:
rather than the original neutral PIP component:
In more complex molecules all piperidine rings will be charged like this.
Trialkylamine to trialkylammonium¶
A SMARTS pattern [$([NX3]([CX4])([CX4])[CX4])] is used to detect nitrogen atoms
with no hydrogen atom attached that are also bonded to three SP3 carbon atoms.
A proton hydrogen atom is added in these cases.
For example, running grade2 for the PDB component
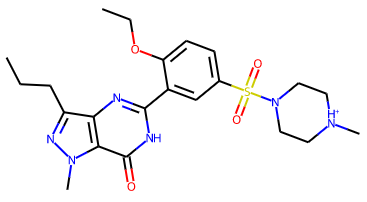
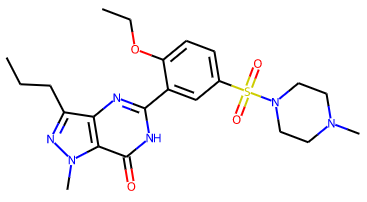
VIA https://www.rcsb.org/ligand/VIA Sildenafil:
$ grade2 --PDB_ligand VIA
Produces the warning messages:
WARNING: Charging groups likely to be charged at neutral pH.
WARNING: ---- If you do not want this, rerun with the option: -N, --no_charging
charging trialkylamine to trialkylammonium
add new proton HN17 onto atom N17 (existing hydrogen atom_ids: None)
Notice that the atom name of the proton hydrogen added is based on the atom name of the nitrogen. The resulting molecule is a cation:
rather than the original neutral VIA component:
Notice that the other piperazine nitrogen atom is not protonated as it is attached to a sulfur atom. The small molecule structure for Sildenafil citrate monohydrate (Yathirajan et al., 2005) CSD entry: FEDTEO shows that Grade2 protonates the correct nitrogen atom.
Charging multiple groups¶
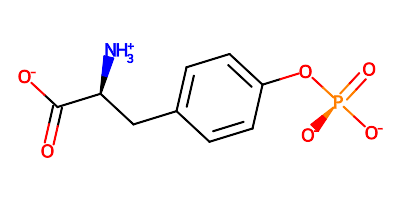
If the input molecule matches more than one of the charging patterns above then each will be applied. For instance, phosphotyrosine https://www.rcsb.org/ligand/PTR will be output with the amino acid in a zwitterionic with phosphate having two negative charges:
The warning messages show that in this case show that 3 charging patterns are applied:
WARNING: Charging groups likely to be charged at neutral pH.
WARNING: ---- If you do not want this, rerun with the option: -N, --no_charging
charging phosphoric acid(s) to phosphate ion(s)
charging carboxylic acid(s) to carboxylate ion(s)
charging alkylamine to alkylammonium
remove proton HXT and add negative charge to atom OXT
remove proton HO2P and add negative charge to atom O2P
remove proton HO3P and add negative charge to atom O3P
add new proton HN3 onto atom N (existing hydrogen atom_ids: H, HN2)