
| Attachments | |
|---|---|
| 2a0f_znb_fix_no_rest.png | 146K |
| 2a0f_znb_orig.png | 133K |
| 2a0f_nohoh_005_autoncs_break_SSBOND_metal_restraint | 1K |
| 2a0f_znd_fix_rest.png | 153K |
| 2a0f_znb_fix_rest.png | 140K |
These are preliminary notes on an initial examination in January 2008. They need to be worked up into a proper tutorial.
2a0f seems a likely target with problematic zinc sites
| run | Rwork/Rfree | Molprobity %Rama Out/score |
| initial pdb | 0.250/0.250(1) | 8.96%/4.39 |
| 2a0f_001_control.log | 0.2236/0.3269 | 11.39%/4.70 |
| 2a0f_002_autoncs.log | 0.2743/0.2966 | 5.86%/4.39 |
| 2a0f_003_autoncs_target1d09.log | 0.2843/0.2881 | 2.43%/3.75 |
| 2a0f_004_autoncs_break_SSBOND.log | 0.2521/0.3302 | 5.97%/4.53 |
| 2a0f_005_autoncs_break_SSBOND_metal_restraints.log | 0.2563/0.3259 | 6.42%/4.50 |
| 2a0f_006_autoncs_target1d09_break_SSBOND.log | 0.2785/0.2970 | 2.32%/3.62 |
| 2a0f_007_autoncs_target1d09_break_SSBOND_metal_restraints | 0.2785/0.2964 | 2.31%/3.61 |
(1) no free set in data
| run | final X-ray weight |
| 2a0f_001 | 1234 |
| 2a0f_002 | 738 |
| 2a0f_003 | 100 |
| 2a0f_004 | 2025 |
| 2a0f_005 | 1620 |
| 2a0f_006 | 184 |
| 2a0f_007 | 184 |
for the metal sorting example B site is mucked up by W296.
| run | Rwork/Rfree | Molprobity %Rama Out/score |
| initial pdb | 0.258/0.263(1) | 8.96%/4.39 |
| 2a0f_nohoh_001_control | 0.2345/0.3413 | 13.94%/4.81 |
| 2a0f_nohoh_002_autoncs | 0.2532/0.3239 | 7.52%/4.55 |
| 2a0f_nohoh_003_autoncs_target1d09 | 0.2769/0.2894 | 2.10%/3.71 |
| 2a0f_nohoh_004_autoncs_break_SSBOND | 0.2501/0.3351 | 10.51%/4.70 |
| 2a0f_nohoh_005_autoncs_break_SSBOND_metal_restraint | 0.2516/0.3338 | 10.84%/4.68 |
| 2a0f_nohoh_006_autoncs_target1d09_break_SSBOND | 0.2686/0.2981 | 2.88%/3.73 |
| 2a0f_nohoh_007_autoncs_target1d09_break_SSBOND_metal_restraints | 0.2635/0.3027 | 2.32%/3.52 |
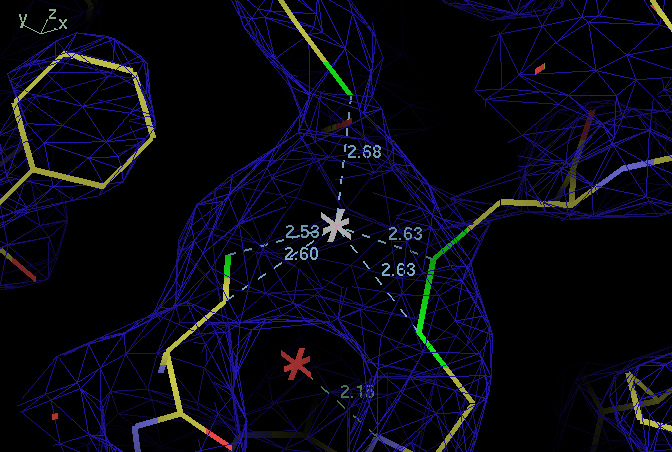
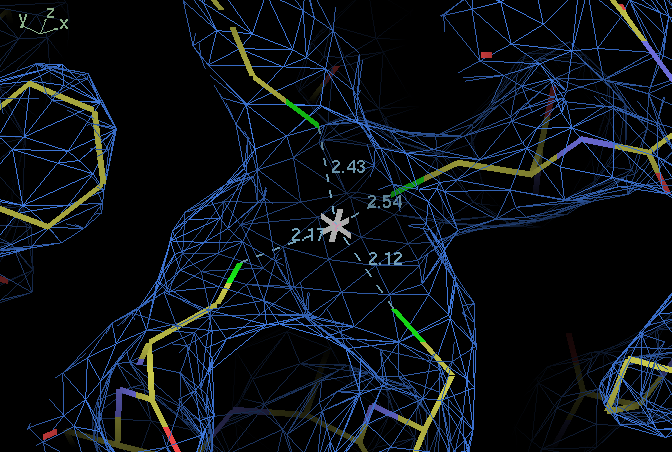
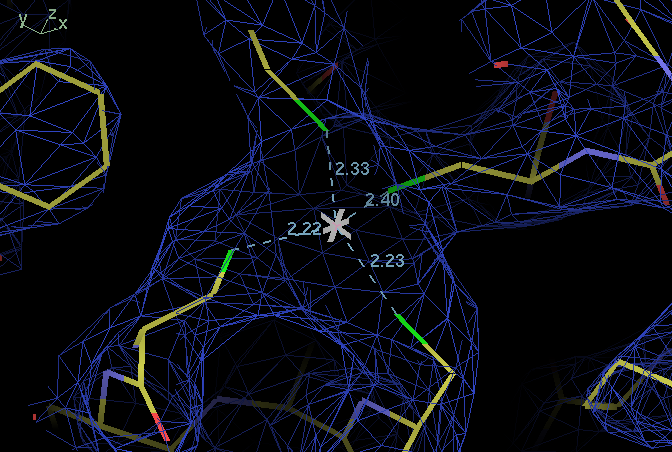
| ligand | B114 SG | B138 SG | B141 SG |
| B109 SG | 113,110 | 112,108 | 118,115 |
| B114 SG | 95,98 | 107,108 | |
| B138 SG | 111,116 |
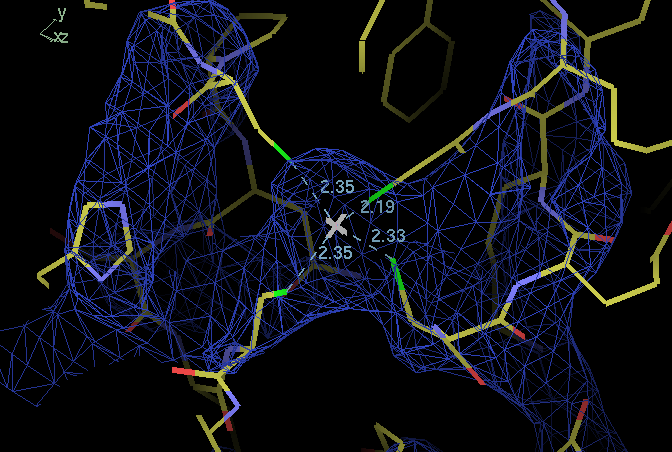
(Impression of 2a0f is that A chain is well placed this is next to ZN binding domain from B chain - reasonable but as one works along C chain and D chain things get worse and worse. Chains are not placed at all reasonably.) Maybe worth looking to see whether it is possible to do properly someday.
Page by Oliver Smart original version 22 June 2008. Address problems, corrections and clarifications buster-develop@globalphasing.com