autoBUSTER Example Application: using NCS in the refinement of a high resolution structure
- chymotrypsin was one of the first enzyme structures to be determined by Blow and coworkers.
- A high resolution structure of the apo enzyme was determined in 1985 and deposited as 4cha. It was refined with the least squares program PROLSQ
- details of 4cha: http://www.ebi.ac.uk/msd-srv/msdlite/atlas/summary/4cha.html
- We will use this as an example to see whether LSSR NCS restraints can help with refinement of high resolution structures.
- In addition it provides a good example showing how water molecules can be included into NCS restraints.
- The example also shows the potential to elucidate useful information by revisiting old structures.
Starting files
- 4cha_with_shell_free.mtz deposited structures factors with a Rfree selection added by the ccp4 program sftools. As 4cha has NCS the selection of the set was done in thin shells with the command
RFREE 0.05 SHELL
autoNCS can help a high resolution structure
- 4chaNCS_001_control_noncs provides a control autoBUSTER without restraints on the NCS.
- Default is not to do any water insertion, just keep waters as the provided in the input (here those placed in 4cha).
- 4chaNCS_002_autoncs uses automated NCS setup procedure. The additional options used are:
- -autoncs automated NCS procedure is used. This will identify NCS and use the new LSSR restraints on positions. No restraints are applied to temperature factors. The -autoncs procedure includes a "prune out" procedure.
- -sim_swap_equiv this will try to swap equivalent atoms in residues such as PHE and ASP to improve the NCS agreement between the copies.
- 4chaNCS_003_autoncs_weight_0.5 This job checks the result of reducing the weight for LSSR on NCS. The -autoncs_weight 0.5 command line option halves the weight from the default.
- 4chaNCS_004_autoncs_weight_2.0 Checks the effect of doubling the NCS weight.
- Comparing the initial runs in terms of refinement statistics and geometry quality measures (calculated with MolProbity)
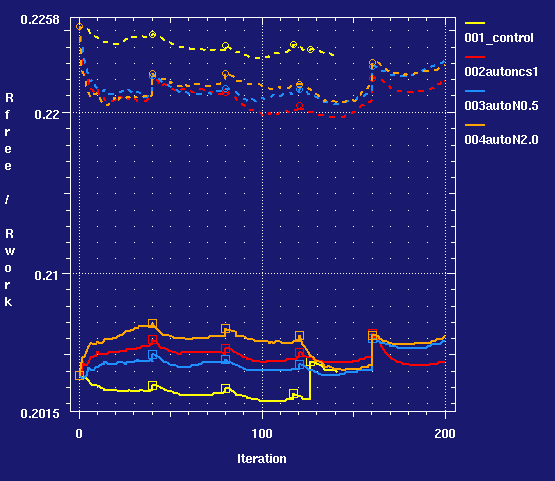
- we can conclude from this that using LSSR restraints on NCS helps improve Rfree, reduce the Rfree-Rwork gap and improve geometry quality measures. The improvements are small but real - it is worthwhile to use NCS at 1.68Å resolution (at least with this geometry function).
- In this case the default weight is good. Results do not vary that much with weight used.
- (002) looking at the structure map there is a lot of difference density due to solvent. To check that NCS has not pulled residues whose conformations actually differ between the two chains out of density, look at the 'LSSR restraints for NCS' section in the LIST.html file in cycle 5: you see
LSSR restraints for NCS
Atoms that are pulled the hardest by LSSR NCS restraints:
(Consider whether side chain or residue NCS partners could be close yet
distinct. If so you may want to prune this out from the relevant NCS set.)
LSSR_grad_norm/ atom
avg(LSSR_grad_norm)
5.27 A|172:NE1 (TRP)
5.23 B|172:NE1 (TRP)
4.57 B|119:CB (SER)
4.51 A|119:CB (SER)
4.28 A|172:CD1 (TRP)
4.26 B|172:CD1 (TRP)
4.16 B|94:O (TYR)
4.15 A|94:O (TYR)
and if you look around those residues in coot there are no difference-map features.
Water Insertion
- autobuster water insertion can be used to progress the model interpretation from the final 002 result.
- It works very well. The final file has 363 water molecules, compared to the 85 in the original 4cha.pdb. The refinement statistics are improved, and Rfree drops by 2.3% with only a small increase in the Rfree, Rwork gap.
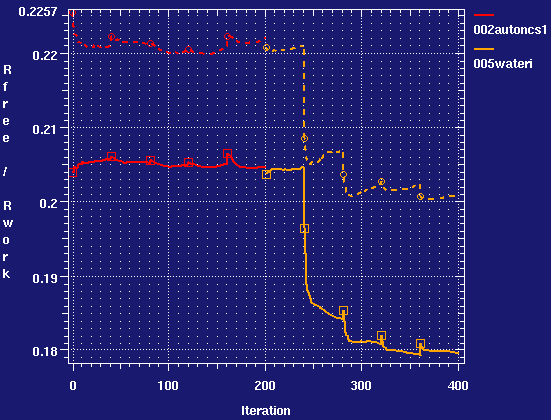
Including water in NCS restraints
- To include water into NCS restraints it is necessary to work out which chain each water molecule is nearest, and to give matching water molecules in the two NCS copies matching residue numbers. This is exactly the function of the ccp4 program sortwater (http://www.ccp4.ac.uk/dist/html/sortwater.html)
- 4chaNCS_006_sortwater uses sortwater to assign the waters from the result of the 005 run into two chains U and V, corresponding to the the proteins chains A and B.
- The sortwater program requires NCS operators. NCS operators determined by a robust but not particularly sophisticated method are reported in the LIST.html file in lines of the form
TRANS_AB (twelve numbers)
TRANS_BA (twelve numbers)
and what you need for sortwater are the numbers after TRANS_BA, and TRANS_CA, TRANS_DA ... if there are more than two chains.
The stand alone programs lsqkab or lsqman can be used to get NCS operators, and have many clever options for obtaining good operators where the similarity between chains is not self-evident.
- sortwater takes the 345 waters in its input file, and works out that they consist of 118 pairs of waters related by NCS together with 109 waters that appear on only one chain.
- the result of the procedure is the coordinate file 4chaNCS_006_sortwater.pdb
- 4chaNCS_007_include_water_in_NCS
- now that the water molecules have been marked up with U and V chain IDs, they can be included into NCS.
- To do this it is necessary to switch from -autoncs to a manual NCS setup using a -Gelly file.
- The easiest way to do this is to start from the file auto_gelly_cards.txt produced in the final cycle of the 005 run:
cat ab_runs/4chaNCS_005_followon_autoncs_waterinsert_result/01-BUSTER/Cycle-5/shell.01/auto_gelly_cards.txt
# cards produced by -autoncs
NOTE BUSTER_SET ncsautoXcld = Water
NOTE BUSTER_SET ncsautoXcld = ncsautoXcld + { A|10 A|20 A|129 A|156 A|163 }
NOTE BUSTER_SET ncsautoXcld = ncsautoXcld + { A|182 A|183 A|184 A|185 A|225 }
NOTE BUSTER_SET ncsautoXcld = ncsautoXcld + { A|226 A|ENDB }
NOTE BUSTER_SET ncsautoABset = Chain_A \ ncsautoXcld
NOTE BUSTER_SIM_DEFINE ncsautoAB ncsautoABset B
NOTE BUSTER_SIM_RESTRAIN_LSSR ncsautoAB
- This provides a basis for the definitions 4chaNCS_007_include_water_in_NCS. The NOTE BUSTER_SIM_JOINT card is used to join the AB set with the UV. This means that the matched water molecules are restrained by LSSR to have similar contacts to the A and B chains of the protein.
- So including water into the NCS results in a very slight (0.1%) reduction in Rfree, and an improvement in Rfree-Rwork gap. At low resolutions the improvement may be bigger.
Implications for 4cha structure
- After this refinement the next appropriate stage is to use coot to fix up the structure. This is outside the area of this tutorial but it is interesting to examine the density around residue B|10. Three extra residues that are missing in 4cha can easily be placed:
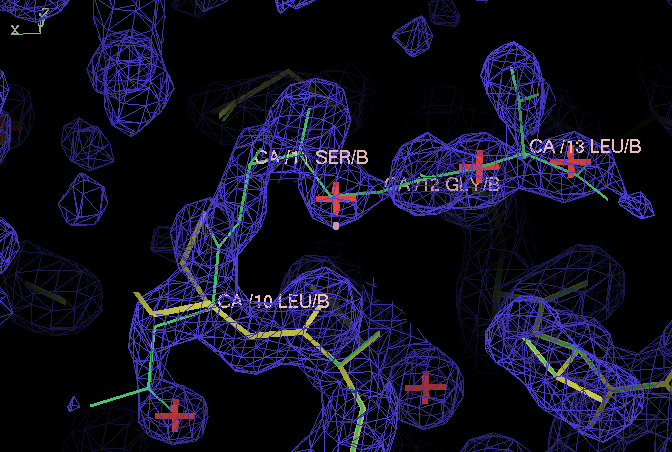 The density is from the 007 run 2Fo-Fc contoured at 1 sigma. The model in thick yellow lines with bold red waters is the final model from the 007 run; the green model shows a placement of the missing residues into density.
The density is from the 007 run 2Fo-Fc contoured at 1 sigma. The model in thick yellow lines with bold red waters is the final model from the 007 run; the green model shows a placement of the missing residues into density.
Page by Oliver Smart original version 21 May 2008. Address problems, corrections and clarifications buster-develop@globalphasing.com



 The density is from the 007 run 2Fo-Fc contoured at 1 sigma. The model in thick yellow lines with bold red waters is the final model from the 007 run; the green model shows a placement of the missing residues into density.
The density is from the 007 run 2Fo-Fc contoured at 1 sigma. The model in thick yellow lines with bold red waters is the final model from the 007 run; the green model shows a placement of the missing residues into density.