Please be aware that CSD 2021 is currently unsupported on MacOS 11 (Big Sur) and therefore Grade (grade and grade_PDB_Ligand) as well as buster-report are not supported on that platform. Support for MacOS 11 is expected in a forthcoming update to CSD.
We've had reports of long execution times for previous releases of BUSTER (or Pipedream) on CentOS8 due to our distributed Gnuplot binary taking a very long time. We can partly reproduce this (although not quite to the same extent), but hope that the binary distributed with this release will on CentOS8 behave similar to other Linux distributions.
If you still experience problems, we would recommend the installation of an OS-specific version of Gnuplot on those systems.
As root, run
yum install gnuplot
and as software installer run
echo "setenv BDG_TOOL_GNUPLOT \"/usr/bin/gnuplot\"" >> $BDG_home/setup_local.csh echo "export BDG_TOOL_GNUPLOT=\"/usr/bin/gnuplot\"" >> $BDG_home/setup_local.sh
The setting of the environment variable BDG_TOOL_GNUPLOT will ensure the use of the newly installed OS-version of "gnuplot".
Remember that CentOS8 will reach end-of-life on December 31, 2021, while CentOS7 is still supported til June 30th, 2024!
The latest Coot version 0.9.6 (e.g. distributed as part of the 7.1.016 update of CCP4) stops with an error when running visualise-geometry-coot: this is due to changes in Coot that removed the no-coot-tips function.
You can fix that by removing the corresponding line 11 in $BDG_home/scripts/problem-gui.scheme, e.g. using
sed -i "s/^(no-coot-tips)//g" $BDG_home/scripts/problem-gui.scheme(or any editor of your choice).
The -onlyRING option to aB_fuseplanes misbehaves on MacOS systems and will most likely show no effect (i.e. fused planes). It works as expected on Linux.
The -cleanTOR option should be avoided with Grade2 dictionaries, since it might drop specific 4-atom restraints by mistake.
When querying the CSD (via Mogul) as part of the buster-report analysis, we are currently converting the compound PDB structure into MOL2 format using Openbabel. As an alternative, the original method of giving the PDB format as-is to Mogul can be selected (by setting the environment variable BDG_MOGUL_USE_PDB to a non-empty value).
This change in the 20201211 release was triggered by some very poor results for particular compounds - mainly due to incorrect determination of bond and atom typing when pure PDB information was given to Mogul. The conversion to MOL2 seems to do a bit better as can be seen here:
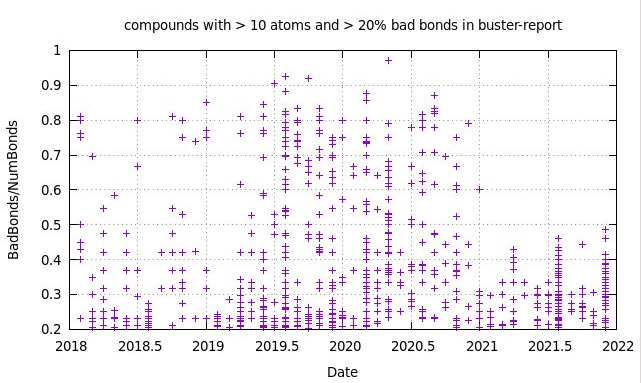 |
We are now avoiding some massively incorrect analysis (where nearly all bonds were marked as bad when using the PDB representation before the 20201211 release). However, what is also obvious is that there are still problems with this approach - not surprisingly, since we know that (1) the bond type assignment in Openbabel can be problematic and (2) there are some specific requirements regarding that assignment when it comes to the CSD analysis itself (see e.g. Appendix B here).
We are working on a much cleaner and more rigorous method of providing exact information about the compound to the CSD analysis step within buster-report. This will be based on the methods and functionality provided by Grade2 and should ensure that (1) the same information as used as restraints is used, and (2) the specific expectations of the Mogul analysis are taken care of. Hopefully, these improvements should become available in the next release.