autoBUSTER Example Application: Using target restraints for 2e4y
Background
- LSSR target restraints enable the exploitation of the similarity of the structure under refinement to an already known structure.
- In this example we will use target restraints to help with the refinement of a low resolution ligand-bound molecular replacement structure.
Starting files
Runs
- We start with a run exploiting both NCS and target similarity. 2e4yTargetExample_001_autoncs_target Options used in script:
- explanation of shell scripts used here
- The -autoncs_noprune option is appropriate to apply NCS for an initial refinement.
- The -target option is set to target the initial molecular replacement solution (the starting point refinement). As this is based on the higher resolution structure 2e4u this is sensible.
- -nbig 10 use 10 big cycles of refinement. For the initial refinement from a molecular replacement solution, it is wise to from the default of 5 big cycles to allow the optimizer to do as much as possible. Subsequent runs after for instance moving some residues in coot could use fewer big cycles, two is often sufficent.
- -RB to do a single cycle of rigid body refinement. This is sensible after molecular replacement, see BUSTER FAQ
- To see the effect of target restraints a series of control runs with different conditions can be performed:
- 2e4yTargetExample_002_control_no_sim
- provides a control where the same conditions are used as the 001 run except that neither NCS nor target restraints are used.
- 2e4yTargetExample_003_control_autoncs
- provides a control where the same conditions are used as the 001 run except that target restraints are not used.
- 2e4yTargetExample_004_control_rigid
- This provides a control where the rigid body definitions are used to keep the protein chains A and B rigid.
- Once again individual temperature factors are allowed to vary subject to tight harmonic B correlation restraints.
- This tests whether allowing the protein to move at all from the higher resolution structure improves the improvement to experimental data.
- Note that the X-ray weight is fixed to that found in the 002 control run - rms bond lengths cannot be to gauge the balance between geometric and X-ray function contributions as the structure is rigid.
Results
- Compare the target run with the controls in terms of raw refinement statistics (updated 2009 better geometry):
(A) pdb quoted values (not autobuster)
- comparative progress graph for runs:
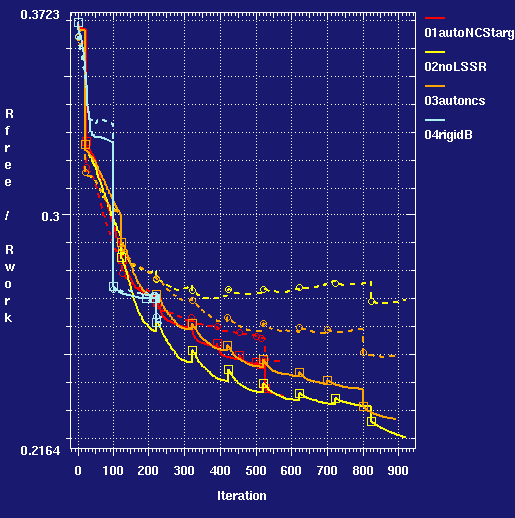
- Using the target restraints produces a lower Rfree than any of the controls. The MolProbity geometry scores are improved from the values for the high resolution structure used as the MR search model.
- Comparing the target restraints run (001) to the control (004) where the protein chain were kept rigid at the higher resolution structure shows that allowing positional flexibility under restraints produces an improvement: Rfree is lower by 1.6%.
- The control run without any similarity restraints (002) produces a slight worsening of geometrical health indicators and a 5% Rfree Rwork gap. Although difference density is still found, the refinement would limit what rebuilding could be done. The actual 2e4y pdb entry structure appears to have been treated in this way, although the geometrical health indices are now slightly better with BUSTER.
- Using NCS helps matters (compare 002 with 003). At low resolution NCS really helps.
- But the combination of NCS and targeting does best (001 is better than 003).
- Looking at the structure and density for the target run (001 run).
- The structure is at 3.40Å resolution so the map is what is to be expected - a tube like backbone with only occasional visible side chains.
- It is useful to look for density features by superposition of the higher resolution structure pdb entry 2e4u (this provided the MR search model).
- There is density in the A copy active site.
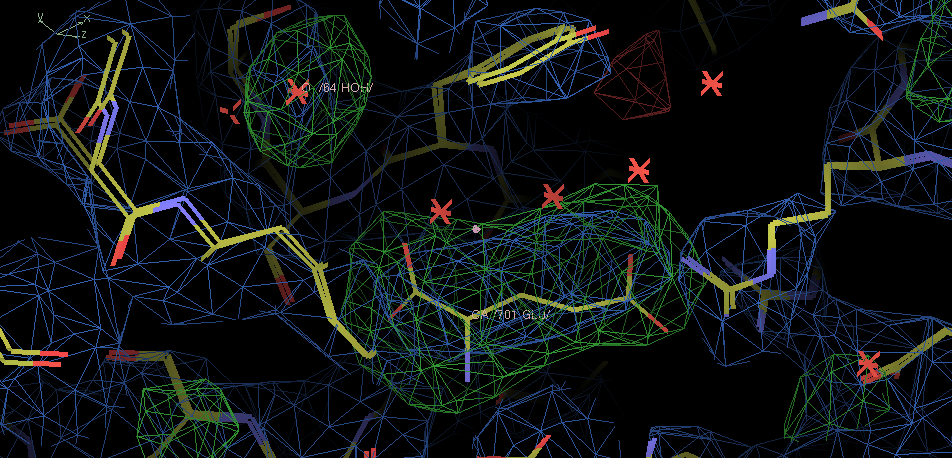 The ligand is larger that the glutamic acid from 2e4u, but this density is not good enough to place it credibly
The ligand is larger that the glutamic acid from 2e4u, but this density is not good enough to place it credibly
- The small C terminal domain and NAG residue chopped out from the MR search model have clear density in the A chain copy:
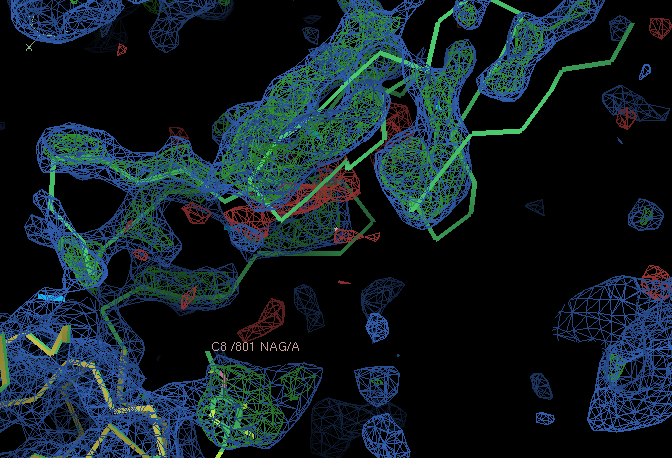
- However, the situation in the B chain is worse. The left out parts only have weak density (the B chain small C terminal domain is not modelled in the deposited structure). The density for one of the domains in the B chain is weak and broken. This would need further exploration to take the structure determination further.
Page by Oliver Smart original version 21 May 2008. Updated July 2009. Address problems, corrections and clarifications to buster-develop@globalphasing.com


 The ligand is larger that the glutamic acid from 2e4u, but this density is not good enough to place it credibly
The ligand is larger that the glutamic acid from 2e4u, but this density is not good enough to place it credibly