
| Attachments | |
|---|---|
| run_autoSHARP_2b.png | 247K |
| run_autoSHARP_1.png | 236K |
| run_autoSHARP_3.png | 97K |
| run_autoSHARP_4.png | 40K |
| autosharp_reference_card.pdf | 218K |
| run_autoSHARP_2a.png | 203K |
| autoproc_reference_card.pdf | 226K |
See also:
If you want to install the software on your own computer(s) (Linux or MacOS): SHARP/autoSHARP home page - free for academics.
We are going to follow one of the examples given here. In case you have trouble downloading the data files from our webpage, you should also be able to copy them from DLS computers:
module load ccp4-workshop
clusterme
module load ccp4-workshop
mkdir -p SHARP
cd SHARP
cp /dls/i03/data/2020/mx27960-4/processing/hcf00057/Workshop/SHARP/* .
ls -l
A very quick example (from the larger list of examples) would be 1O22:
run_autoSHARP.sh \ -fast -nowarp \ -seq 1o22.pir -ha "Se" \ -wvl 0.9778 peak -7 5 -sca 1o22_peak.sca \ -d autoSHARP_SAD-1 | tee autoSHARP_SAD-1.lis
You should then see something like this:
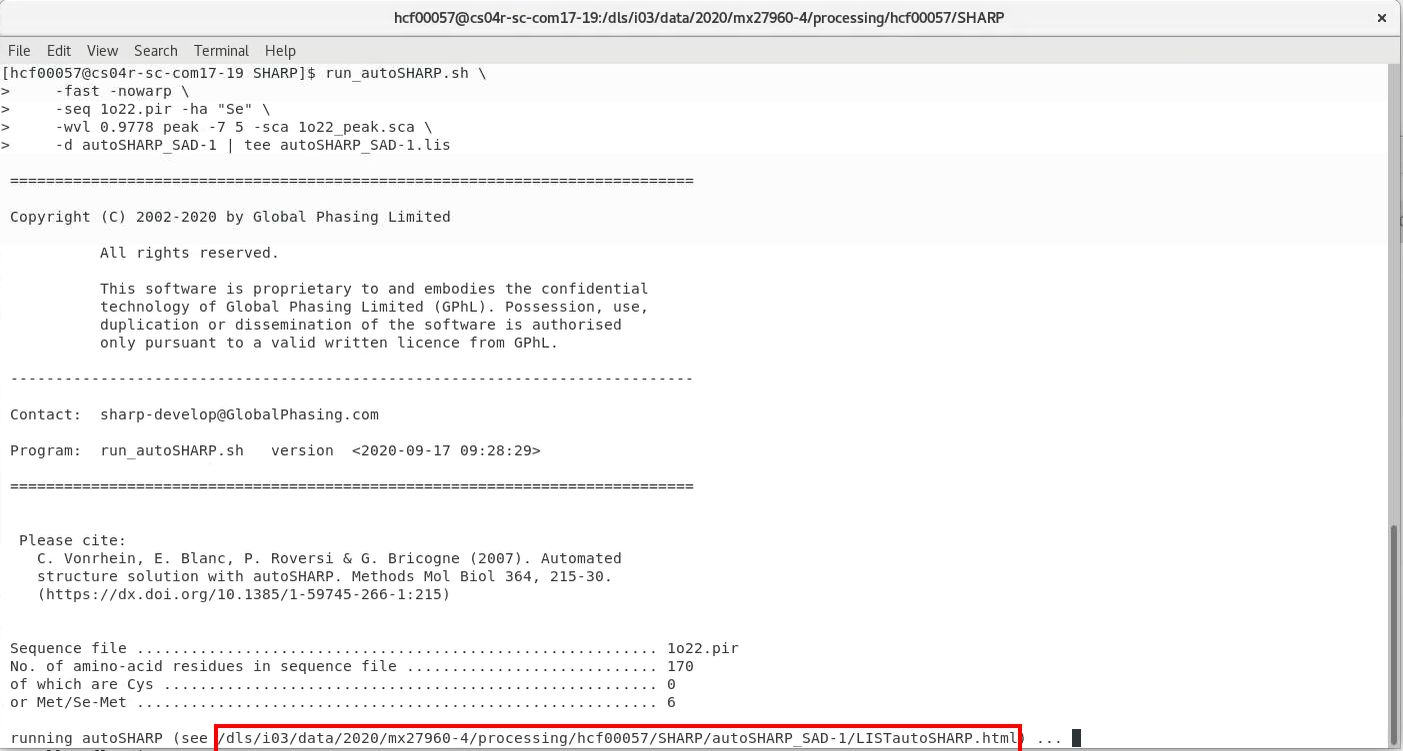 |
pointing you to the output file (a HTML file with the name LISTautoSHARP.html). Please open this file in a browser (e.g. Firefox) to see the full autoSHARP log file and results. You will some results regarding data analysis and substructure solution (with SHELXC/D):
 |
and HA refinement and phasing (with SHARP), density modification (SOLOMON) and model-building (PARROT+BUCCANEER):
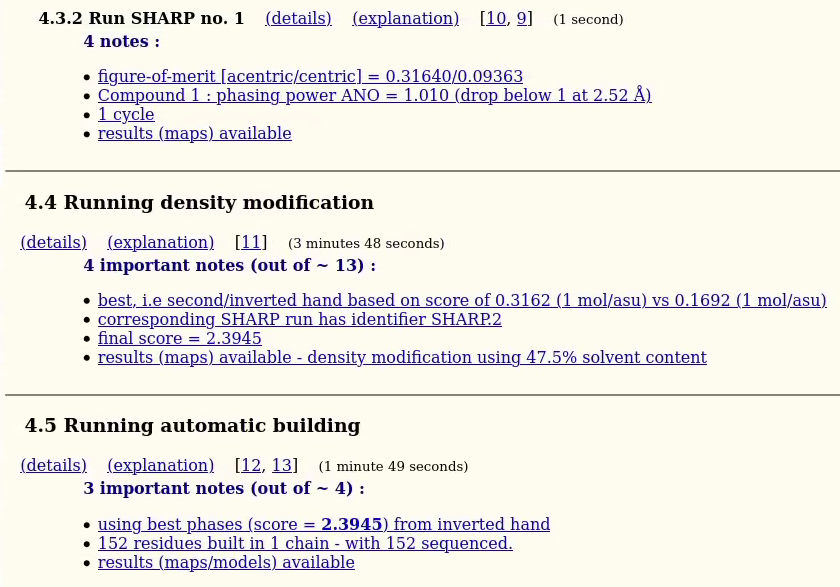 |
Of course, this is a nice simple SAD example that runs very fast:
 |
(it is nearly always the density-modification and building stage that takes the vast amount of running time).
At various times the HTML file will mention results being available - eg.
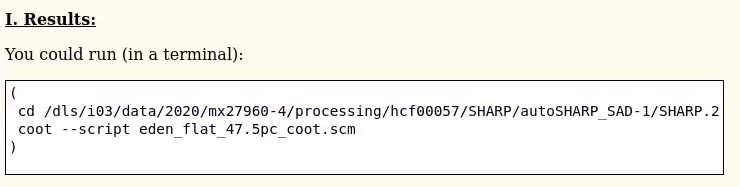 |
You can cut-n-paste the text from the white box (including opening and closing brackets!) into a terminal window. At the end of the run, you will also see a summary on standard output (i.e. the Terminal where you executed the run_autoSHARP.sh command).
If you want to use some of the Thaumatin datasets collected on Monday:
wget -O Thaumatin.seq https://www.rcsb.org/fasta/entry/1RQWmodule load ccp4-workshop clusterme module load ccp4-workshop mkdir -p SHARP cd SHARP run_autoSHARP.sh \ -seq Thaumatin.seq \ -ha Se -nsit 4 \ -wvl 0.97927 peak -6.47 5.17 \ -mtz /dls/i04/data/2020/mx27960-1/processed/Test_Thaumatin/Thau1/thauMAD_pk_1_/autoPROC/staraniso_alldata-unique.mtz \ -R 50.0 2.0 -fast -nowarp \ -d thauMAD_pk_1 | tee thauMAD_pk_1.lis
Although we would always recommend to start with a run of autoSHARP using all defaults, for these workshop examples we are going to use a few extra flags:
run_autoSHARP.sh -nowarp ...Although ARP/wARP building would potentially result in a much better built model (not necessary more complete), it doesn't scale with multiple processors/threads and therefore would overrun the tutorial time. You could also use
run_autoSHARP.sh -nowarp -fast ...or even
run_autoSHARP.sh -nobuild -fast ...if you are only interested in experimental phasing up to the first (hopefully interpretable) electron density map.
At the end you should be presented (on standard output, i.e. within the Terminal) with something like
RESULTS:
[01] HA detection (SHELXC/D) :
initial sites = MAD_3ISY.01/SHELX/1.1.hatom
[02] HA refinement and phasing (SHARP, first run) :
refined sites = MAD_3ISY.01/SHARP.1/hatom.pdb
phases = MAD_3ISY.01/SHARP.1/eden.mtz
residual maps = MAD_3ISY.01/SHARP.1/resid.mtz
[03] HA refinement and phasing (SHARP, final run) :
refined sites = MAD_3ISY.01/SHARP.3/hatom.pdb
phases = MAD_3ISY.01/SHARP.3/eden.mtz
residual maps = MAD_3ISY.01/SHARP.3/resid.mtz
eden.mtz: see $SHARP_home/docs/sharp/manual/appendix1.html#eden
resid.mtz: see $SHARP_home/docs/sharp/manual/appendix1.html#resid
[04] First density modification (after hand-determination) :
phases = MAD_3ISY.01/SHARP.3/eden_flat_45.1pc.mtz
see $SHARP_home/docs/sharp/manual/appendix1.html#edenflat for details
(use FBshasol/PHIBshasol for map calculation or viewing)
[05] First model (built into initial density-modified map) :
model = MAD_3ISY.01/SHARP.3/LJS_45.1pc/buccaneer_cycle-5.pdb
phases = MAD_3ISY.01/SHARP.3/LJS_45.1pc/parrot_cycle-5.mtz
(use parrot.F_phi.F/parrot.F_phi.phi for map calculation or viewing)
[06] Final density modification (after inclusion of previously built models) :
phases = MAD_3ISY.01/SHARP.3/eden_flat_44.8pc.mtz
see $SHARP_home/docs/sharp/manual/appendix1.html#edenflat for details
(use FBshasol/PHIBshasol for map calculation or viewing)
[07] Final model (built into last density-modified map) :
model = MAD_3ISY.01/SHARP.3/LJS_44.8pc/buccaneer_cycle-5.pdb
phases = MAD_3ISY.01/SHARP.3/LJS_44.8pc/parrot_cycle-5.mtz
(use parrot.F_phi.F/parrot.F_phi.phi for map calculation or viewing)
NOTE: results are also collected in directory MAD_3ISY.01/Results
-rw-rw----+ 1 hcf00057 mx27960_4 85 Dec 2 13:01 MAD_3ISY.01/Results/01_1.1.hatom
-rw-rw----+ 1 hcf00057 mx27960_4 1014 Dec 2 13:18 MAD_3ISY.01/Results/02_coot.scm
-rw-rw----+ 1 hcf00057 mx27960_4 509268 Dec 2 13:02 MAD_3ISY.01/Results/02_eden.mtz
-rw-rw----+ 1 hcf00057 mx27960_4 366 Dec 2 13:02 MAD_3ISY.01/Results/02_hatom.pdb
-rw-rw----+ 1 hcf00057 mx27960_4 355204 Dec 2 13:02 MAD_3ISY.01/Results/02_resid.mtz
-rw-rw----+ 1 hcf00057 mx27960_4 1014 Dec 2 13:18 MAD_3ISY.01/Results/03_coot.scm
-rw-rw----+ 1 hcf00057 mx27960_4 509268 Dec 2 13:02 MAD_3ISY.01/Results/03_eden.mtz
-rw-rw----+ 1 hcf00057 mx27960_4 366 Dec 2 13:02 MAD_3ISY.01/Results/03_hatom.pdb
-rw-rw----+ 1 hcf00057 mx27960_4 355128 Dec 2 13:02 MAD_3ISY.01/Results/03_resid.mtz
-rw-rw----+ 1 hcf00057 mx27960_4 417 Dec 2 13:18 MAD_3ISY.01/Results/04_coot.scm
-rw-rw----+ 1 hcf00057 mx27960_4 509668 Dec 2 13:08 MAD_3ISY.01/Results/04_eden_flat_45.1pc.mtz
-rw-rw----+ 1 hcf00057 mx27960_4 366 Dec 2 13:02 MAD_3ISY.01/Results/04_hatom.pdb
-rw-rw----+ 1 hcf00057 mx27960_4 78574 Dec 2 13:10 MAD_3ISY.01/Results/05_buccaneer_cycle-5.pdb
-rw-rw----+ 1 hcf00057 mx27960_4 490 Dec 2 13:18 MAD_3ISY.01/Results/05_coot.scm
-rw-rw----+ 1 hcf00057 mx27960_4 1112676 Dec 2 13:09 MAD_3ISY.01/Results/05_parrot_cycle-5.mtz
-rw-rw----+ 1 hcf00057 mx27960_4 417 Dec 2 13:18 MAD_3ISY.01/Results/06_coot.scm
-rw-rw----+ 1 hcf00057 mx27960_4 508968 Dec 2 13:16 MAD_3ISY.01/Results/06_eden_flat_44.8pc.mtz
-rw-rw----+ 1 hcf00057 mx27960_4 79465 Dec 2 13:18 MAD_3ISY.01/Results/07_buccaneer_cycle-5.pdb
-rw-rw----+ 1 hcf00057 mx27960_4 490 Dec 2 13:18 MAD_3ISY.01/Results/07_coot.scm
-rw-rw----+ 1 hcf00057 mx27960_4 1111336 Dec 2 13:18 MAD_3ISY.01/Results/07_parrot_cycle-5.mtz
You should be able to run Coot with the supplied scripts, e.g.:
cd MAD_3ISY.01/Results
coot --script 07_coot.scm
giving you very detailed information about the results (what they represent, how to view them and generally what to do with them). Further down you see a suggestion about refinement:
Files ready for refinement are:
MAD_3ISY.01/Results/model.pdb
MAD_3ISY.01/Results/for_refinement.mtz
e.g. using BUSTER with
refine -p MAD_3ISY.01/Results/model.pdb \
-m MAD_3ISY.01/Results/for_refinement.mtz \
-autoncs \
-d BUSTER.01 | tee BUSTER.01.lis
followed by
coot --pdb BUSTER.01/refine.pdb --auto BUSTER.01/refine.mtz