
From the session website: We often collect data that, for a variety of reasons, contain spots with less-than-ideal shape/properties. This dataset is a good example of a 'typical' membrane protein dataset with a long unit cell axis. Note that the data for this example are organized in three directories representing the three wavelengths used for collection: peak wavlength ('pk'), inflection point ('ip') and high-remote wavelength ('hrem'). All three wavelengths will be necessary for successful phasing. |
Content:
Just running
% find_images
shows 3 datasets:
SK3_2_###.img : 0 - 269 SK3_4_###.img : 0 - 360 SK3_5_###.img : 0 - 360
We have images with an image number zero: these are often direct-beam shots (to get very accurate direct beam position). Let's check:

From that direct-beam shot image, we can read the direct beam centre in pixels as (X,Y)=(1535,1528). Does that fit with the image header information?
% imginfo *001.img
gives
################# File = SK3_2_001.img >>> Image format detected as MARCCD ===== Header information: date = 24 Nov 2006 13:32:00 exposure time [seconds] = 2.000 distance [mm] = 370.000 wavelength [A] = 0.971500 Phi-angle (start, end) [degree] = 0.000 0.500 Oscillation-angle in Phi [degree] = 0.500 Omega-angle [degree] = 0.000 Chi-angle [degree] = 0.000 2-Theta angle [degree] = 0.000 Pixel size in X [mm] = 0.073242 Pixel size in Y [mm] = 0.073242 Number of pixels in X = 3072 Number of pixels in Y = 3072 Beam centre in X [mm] = 113.232 Beam centre in X [pixel] = 1546.000 Beam centre in Y [mm] = 114.088 Beam centre in Y [pixel] = 1557.689 Overload value = 65535 ################# File = SK3_4_001.img >>> Image format detected as MARCCD ===== Header information: date = 24 Nov 2006 15:39:51 exposure time [seconds] = 1.000 distance [mm] = 370.000 wavelength [A] = 0.979000 Phi-angle (start, end) [degree] = 0.000 0.500 Oscillation-angle in Phi [degree] = 0.500 Omega-angle [degree] = 0.000 Chi-angle [degree] = 0.000 2-Theta angle [degree] = 0.000 Pixel size in X [mm] = 0.073242 Pixel size in Y [mm] = 0.073242 Number of pixels in X = 3072 Number of pixels in Y = 3072 Beam centre in X [mm] = 113.232 Beam centre in X [pixel] = 1546.000 Beam centre in Y [mm] = 114.088 Beam centre in Y [pixel] = 1557.689 Overload value = 65535 ################# File = SK3_5_001.img >>> Image format detected as MARCCD ===== Header information: date = 24 Nov 2006 16:15:15 exposure time [seconds] = 1.000 distance [mm] = 370.000 wavelength [A] = 0.979200 Phi-angle (start, end) [degree] = 0.000 0.500 Oscillation-angle in Phi [degree] = 0.500 Omega-angle [degree] = 0.000 Chi-angle [degree] = 0.000 2-Theta angle [degree] = 0.000 Pixel size in X [mm] = 0.073242 Pixel size in Y [mm] = 0.073242 Number of pixels in X = 3072 Number of pixels in Y = 3072 Beam centre in X [mm] = 113.232 Beam centre in X [pixel] = 1546.000 Beam centre in Y [mm] = 114.088 Beam centre in Y [pixel] = 1557.689 Overload value = 65535
Since this is a Se-MET experiment, it looks like
SK3_2_###.img = hrem SK3_4_###.img = peak SK3_5_###.img = infl
In what order were they collected? We run
% imgdate.sh -s *.img > img.lis
to get
# sorted list of: file, Epoch, Date, seconds-to-previous SK3_2_000.img 1164375106 24 Nov 2006 13:31:46 0 SK3_2_001.img 1164375120 24 Nov 2006 13:32:00 14 SK3_2_002.img 1164375124 24 Nov 2006 13:32:04 4 ... SK3_2_269.img 1164375954 24 Nov 2006 13:45:54 3 SK3_4_000.img 1164382776 24 Nov 2006 15:39:36 6822 SK3_4_001.img 1164382791 24 Nov 2006 15:39:51 15 SK3_4_002.img 1164382794 24 Nov 2006 15:39:54 3 ... SK3_4_360.img 1164383908 24 Nov 2006 15:58:28 3 SK3_5_000.img 1164384901 24 Nov 2006 16:15:01 993 SK3_5_001.img 1164384915 24 Nov 2006 16:15:15 14 SK3_5_002.img 1164384918 24 Nov 2006 16:15:18 3
So we have a large gap between the last hrem image (but maybe something went wrong there, since the images stop with 269 and not the 360 images we have for the other wavelengths) and the first peak image. There is also a larger gap (about 15 minutes) between the peak and infl dataset: maybe that is the time it takes to change wavelength (and realign beam) on that beamline - or maybe it is a different crystal or a different part of the same crystal?
It's always a good idea to not just looking at the first image, but a few images at different viewing angles (relative to the first image), eg. +30, +60 and +90 degree:
| Image | Full | Centre region | Upper left |
| SK3_2_001.img |  |
 |
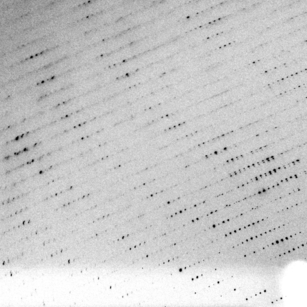 |
| SK3_2_061.img |  |
 |
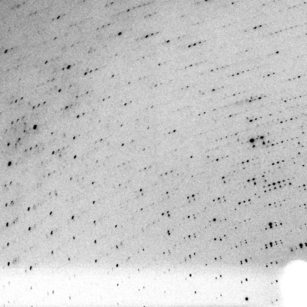 |
| SK3_2_121.img |  |
 |
 |
| SK3_2_181.img |  |
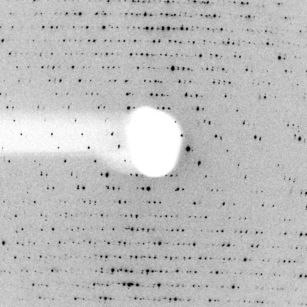 |
 |
The first image shows relative nice lunes, but also already some split (?) spots. This becomes worse with later images.
The first images for all three wavelengths (hrem, peak and infl) show nearly identical diffraction patterns (apart from a slight scaling due to the change in wavelength). So it is most likely that the datasets come from the same crystal - but if they were collected on the same spot of the crystal or on a fresh spot is still unclear.
This looks like a challenging example with possibly multiple lattices - so a slightly more complex approach for running autoPROC:
% process -M automatic -d 01 | tee 01.lis
This will read a so-called macro (named "automatic") to set various parameters which could especially improve the initial indexing stage. To see a list of available (and/or user-defined):
% process -M list
This will give a F222 solution with
Summary data for Project: Test Crystal: A Dataset: 0.971500
Overall InnerShell OuterShell
---------------------------------------------------------------------------
Low resolution limit 68.142 68.142 3.200
High resolution limit 3.189 14.752 3.189
Rmerge 0.103 0.034 0.296
Ranom 0.090 0.025 0.268
Rmeas (within I+/I-) 0.112 0.032 0.347
Rmeas (all I+ & I-) 0.114 0.038 0.345
Rpim (within I+/I-) 0.065 0.019 0.217
Rpim (all I+ & I-) 0.049 0.017 0.172
Total number of observations 332783 1878 1459
Total number unique 62862 408 425
Mean(I)/sd(I) 14.7 37.0 3.8
Completeness 98.1 56.7 68.2
Multiplicity 5.3 4.6 3.4
Anomalous completeness 95.6 55.3 57.8
Anomalous multiplicity 2.8 2.7 1.9
Summary data for Project: Test Crystal: A Dataset: 0.979000
Overall InnerShell OuterShell
---------------------------------------------------------------------------
Low resolution limit 68.142 68.142 3.494
High resolution limit 3.483 14.752 3.483
Rmerge 0.126 0.061 0.222
Ranom 0.111 0.051 0.202
Rmeas (within I+/I-) 0.132 0.060 0.242
Rmeas (all I+ & I-) 0.137 0.067 0.242
Rpim (within I+/I-) 0.070 0.032 0.132
Rpim (all I+ & I-) 0.053 0.028 0.096
Total number of observations 309564 4125 698
Total number unique 47265 697 111
Mean(I)/sd(I) 13.9 29.7 8.4
Completeness 95.7 96.9 22.7
Multiplicity 6.5 5.9 6.3
Anomalous completeness 93.4 95.3 22.6
Anomalous multiplicity 3.4 3.5 3.2
Summary data for Project: Test Crystal: A Dataset: 0.979200
Overall InnerShell OuterShell
---------------------------------------------------------------------------
Low resolution limit 68.142 68.142 3.340
High resolution limit 3.330 14.752 3.330
Rmerge 0.148 0.046 0.312
Ranom 0.137 0.039 0.308
Rmeas (within I+/I-) 0.160 0.046 0.377
Rmeas (all I+ & I-) 0.160 0.051 0.347
Rpim (within I+/I-) 0.083 0.025 0.215
Rpim (all I+ & I-) 0.061 0.021 0.148
Total number of observations 363672 4079 461
Total number unique 54321 696 92
Mean(I)/sd(I) 13.3 36.1 5.6
Completeness 96.4 96.8 17.4
Multiplicity 6.7 5.9 5.0
Anomalous completeness 93.8 93.2 16.7
Anomalous multiplicity 3.5 3.5 2.6
and files:
Can those already be used for solving the structure? See the autoSHARP tutorial.