
| Attachments | |
|---|---|
| 1pmq.pdb | 268K |
| 1pmq.mtz | 512K |
| vis_06.png | 191K |
| vis_01.png | 38K |
| vis_03.png | 214K |
| vis_05.png | 25K |
| 880.grade_PDB_ligand.cif | 16K |
| buster_reference_card.pdf | 171K |
Content:
Remember that you have a BUSTER reference card ({file:buster_referene_card.pdf}) to get help - or search this Wiki (box on the upper left).
In the sections below, the commands you should type into a terminal are shown in green - as in
ls -ltra
The main command you will be using is the command "refine". This comes with inline help via
refine -hwhich will also point you to the locally installed manual.
You can request a licence for BUSTER (free to academics) at https://www.globalphasing.com/buster/.
A good example of using BUSTER (and associated tools) would be 1PMQ.
Create a fresh directory e.g. via
mkdir ~/BUSTER-tutorialand go there with
cd ~/BUSTER-tutorial(or use some other location if you want). You can get the required data files via
fetch_PDB 1PMQ(Note: this will not work at the moment as expected, since 1PMQ was superseded by 4Z9L).
This tool can be used to fetch any model/data from the PDB, resulting in two files:
1PMQ/1pmq.pdb 1PMQ/1pmq.mtz
Or save the following files:
You will also need a restraints dictionary for residue 880 - either from the Grade webserver or save the file
To just compute a map you would use the command (check your BUSTER reference card: buster_reference_card.pdf)
refine -p 1pmq.pdb -m 1pmq.mtz -l 880.grade_PDB_ligand.cif -M MapOnly -d MapOnly | tee MapOnly.lisand then visualise the results using
cd MapOnly visualise-geometry-coot
Alternatively, you could run a full series of BUSTER refinements that are rather typical for getting started with BUSTER on a structure without NCS:
refine -p 1pmq.pdb -m 1pmq.mtz -RB -l 880.grade_PDB_ligand.cif -d 01 | tee 01.lis refine -p 01/refine.pdb -m 1pmq.mtz -M TLSbasic -l 880.grade_PDB_ligand.cif -d 02 | tee 02.lis refine -p 02/refine.pdb -m 1pmq.mtz -M TLSalternate -TLS -M WaterUpdatePkmaps -l 880.grade_PDB_ligand.cif -d 03 | tee 03.lis
The above is just an example of BUSTER refinements when starting from a deposited PDB structure or when switching from a different refinement program to BUSTER. Different starting points (poor initial model, domains missing, low-resolution etc) might need a slightly different approach. See the extensive material on this wiki for some pointers ...
The results of a BUSTER refinement can be loaded into Coot using e.g.
cd 03 coot --pdb refine.pdb --auto refine.mtz
But visualisation might be better done via
cd 03 visualise-geometry-coot
For details about this tool please see here. You will be presented with a box showing you so-called 'unhappy' atoms: these are atoms you can work through to see if there is something that needs manual attention.
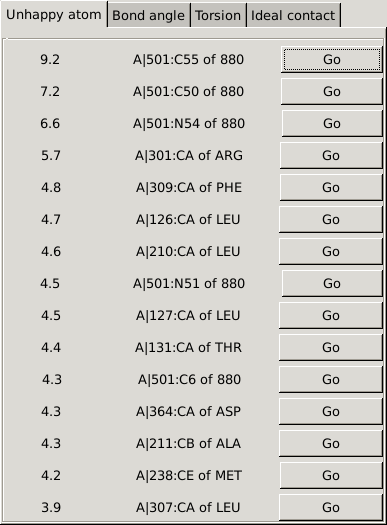 |
Also, all other Coot tools can be used to analyse your structure - which would show you a potential issue at residue A45:
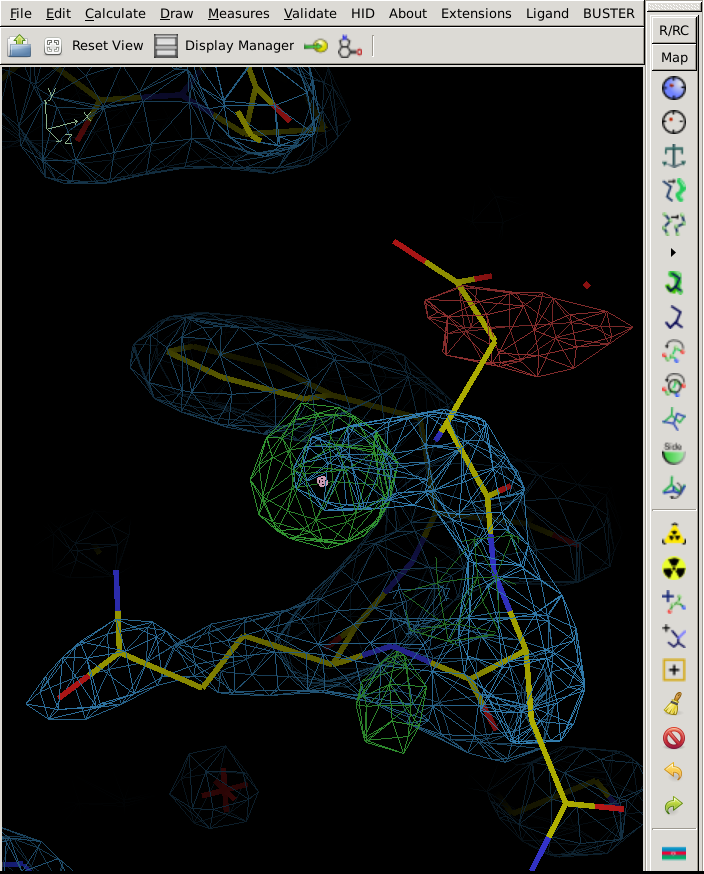 |
After correcting this, you can start a quick BUSTER job using the "BUSTER" button at the top right
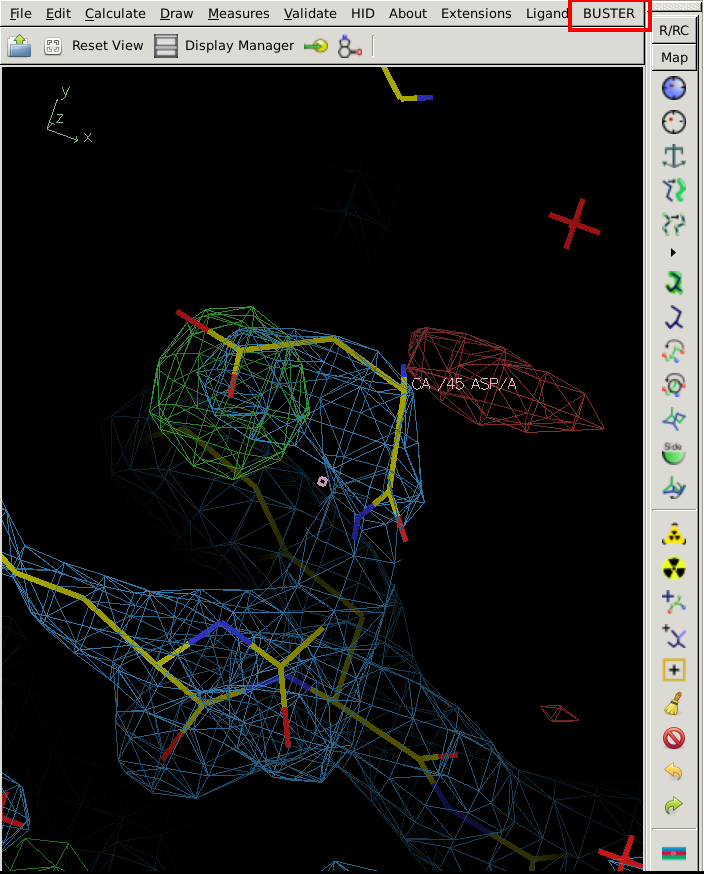 |
After pointing to the correct MTZ file (1pmq.mtz) you can just compute a BUSTER map or do a short refinement:
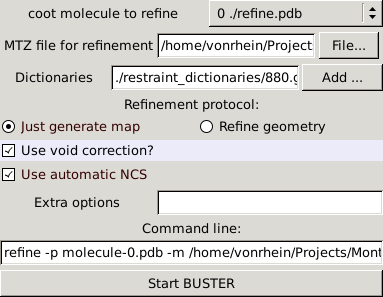 |
Following those initial steps, any subsequent BUSTER refinement would use commands like
refine -p 03/refine-coot-0.pdb -m 1pmq.mtz -M TLSalternate -TLS -l 880.grade_PDB_ligand.cif -d 04 | tee 04.lis
Details:
If you want to work with your own structure: