
| Attachments | |
|---|---|
| 0_eden_flat_53.7pc.mtz.gz | 465K |
| 2QVO-SAD-WikiRun0.0_view1.png | 49K |
| 2QVO-SAD-WikiRun0.0_view2.png | 16K |
| 0_eden_flat_53.7pc_warpNtrace.mtz.gz | 245K |
| 0_eden.mtz.gz | 444K |
| 0_hatom.pdb.gz | 239B |
| 0_eden_flat_53.7pc_warpNtrace.pdb.gz | 14K |
Content
See also the PDB summary.
From an all-defaults autoPROC run we have a highly-redundant 2.3A dataset. Since it was collected at 1.9A wavelength, the idea is to use S-SAD phasing for structure solution.
We also have the sequence (95 residues, 4 Met and 1 Cys):
MEDERIKLLFKEKALEILMTIYYESLGGNDVYIQYIASKVNSPHSYVWLIIKKFEEAKMVECELEGRTKIIRLTDKGQKI AQQIKSIIDIMENDT
We start autoSHARP through the default (httpd-based) interface and give it the following information for a SAD experiment:
which after 10 minutes gives us
1 chain with 87 residues : 1 docked in sequence (87 residues [R/Rfree=0.236/0.353]
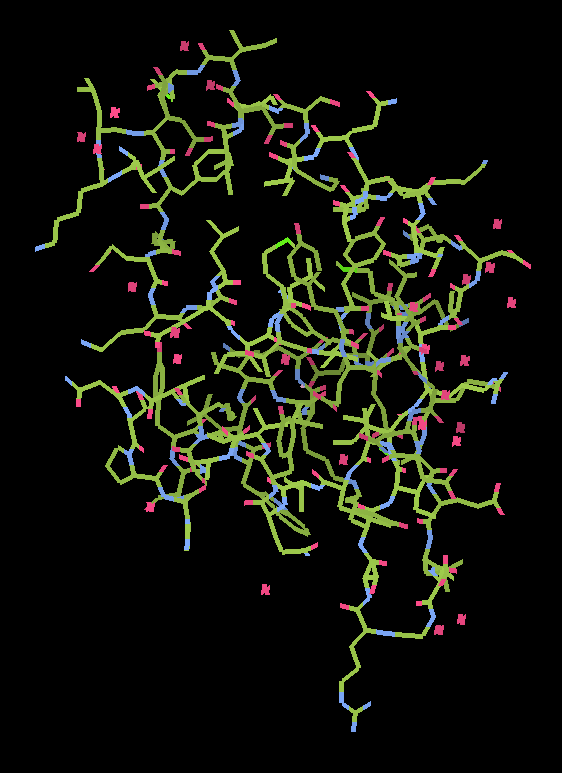 |
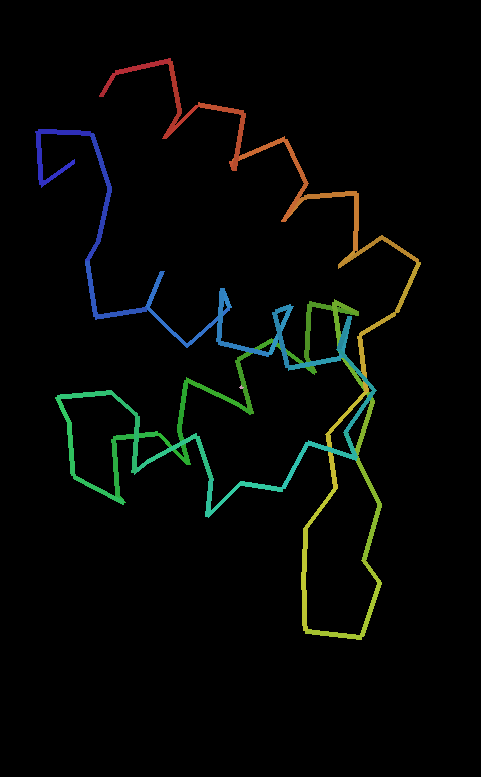 |
Files: